Lecture Four
Proteins & EnzymesDr. Khalidah Merzah
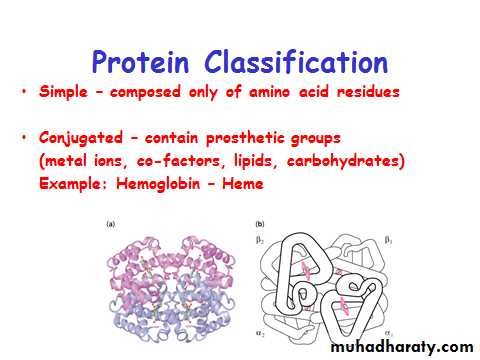
Protein Classification
Protein Classification
One polypeptide chain - monomeric proteinMore than one - multimeric protein
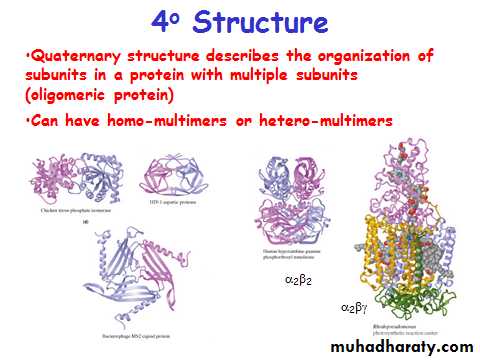
Homomultimer - one kind of chain
Heteromultimer - two or more different chains
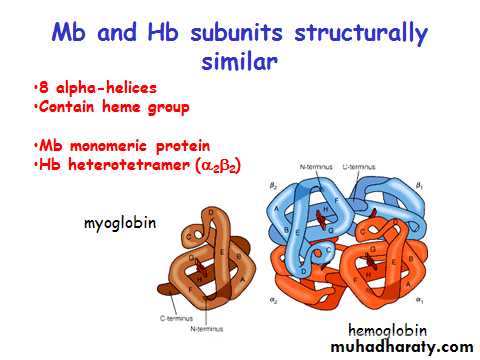
(e.g. Hemoglobin is a heterotetramer. It has two alpha chains and two beta chains.)
Protein Classification
Fibrous –
polypeptides arranged in long strands or sheets
water insoluble (lots of hydrophobic AA’s)
strong but flexible
Structural (keratin, collagen)
Globular –
polypeptide chains folded into spherical or globular form
water soluble
contain several types of secondary structure
diverse functions (enzymes, regulatory proteins)
Protein Function
Catalysis – enzymes
Structural – keratin
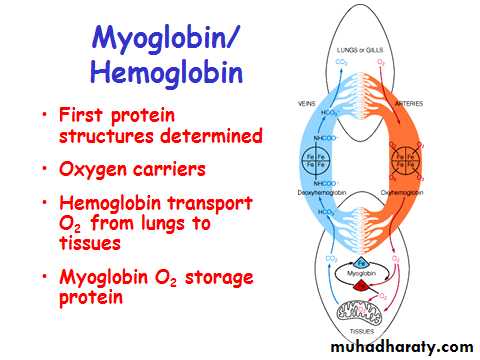
Transport – hemoglobin
Trans-membrane transport – Na+/K+ ATPases
Toxins – rattle snake venom, ricin
Contractile function – actin, myosin
Hormones – insulin
Storage Proteins – seeds and eggs
Defensive proteins – antibodies
Non-covalent forces important in determining protein structure
van der Waals: 0.4 - 4 kJ/molhydrogen bonds: 12-30 kJ/mol
ionic bonds: 20 kJ/mol
hydrophobic interactions: <40 kJ/mol
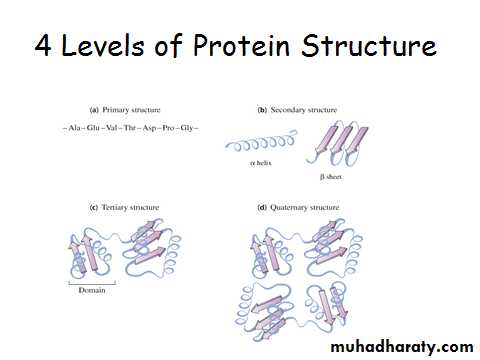
1o Structure Determines 2o, 3o, 4o Structure
Sickle Cell Anemia – single amino acid change in hemoglobin related to disease
Osteoarthritis – single amino acid change in collagen protein causes joint damage
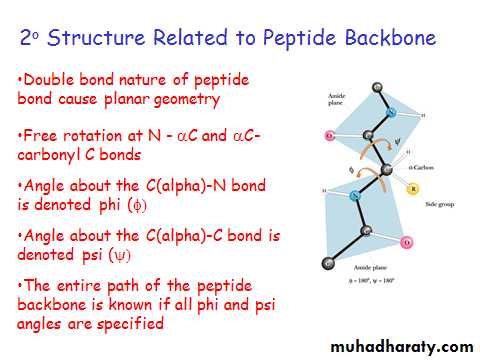
Classes of 2o Structure
Alpha helix
B-sheet
Loops and turns
Protein 3-D structure: 3o and 4o structure and protein folding.
3o Structure●Third level of protein organization
• folding of polypeptide chain causes 2o structures to interact
• Formation of motifs and domains
4o Structure
• Subunits held together by non-covalent interactions
• Oligomeric protein is more stable than disassociated subunits• Active site often made up of AA residues from different subunits
• 4o and 3o structure is often affected by ligand (substrate or inhibitor) binding. Important in enzyme regulationProtein denaturation
●Denaturation – disruption of native conformation
• Heat commonly used to denature proteins
• Tm = temperature where 50% folded/50% unfolded.
• Typical Tm = 40-60oC
• Tm for thermophiles >100oC
(Taq DNA polymerase)
• Chemical denaturants Chaotrophic agents = Urea, KCN detergents = SDS
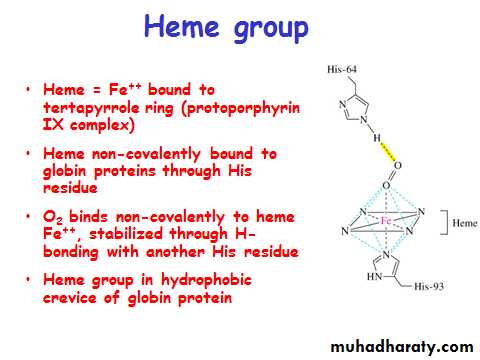
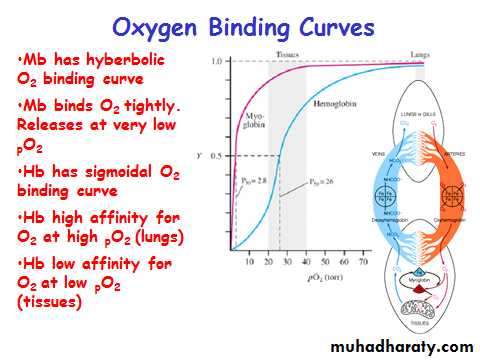
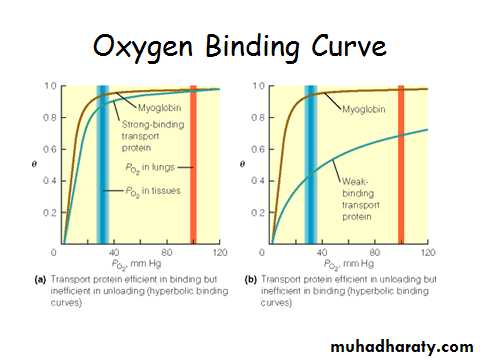
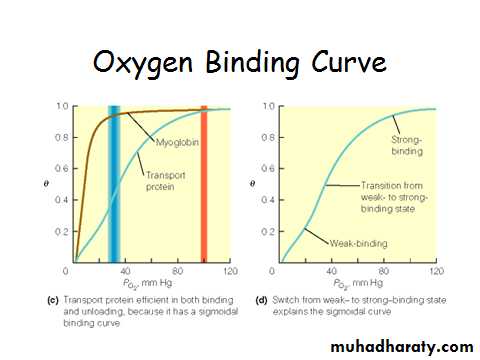
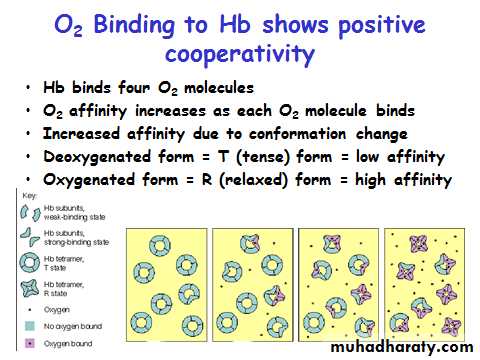
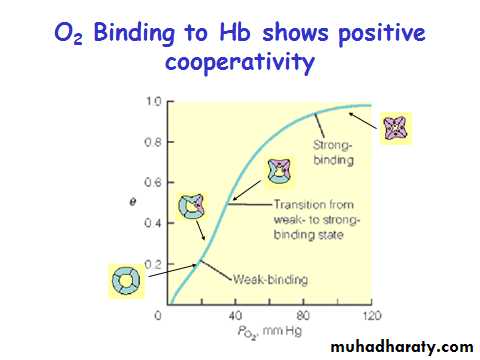
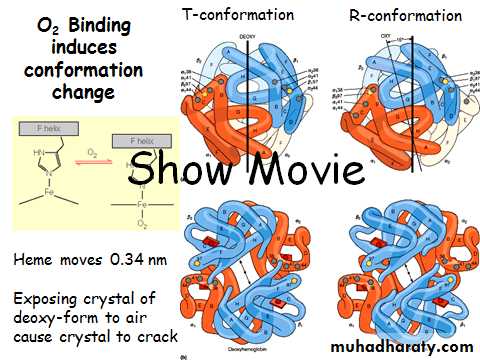
Myoglobin/ Hemoglobin
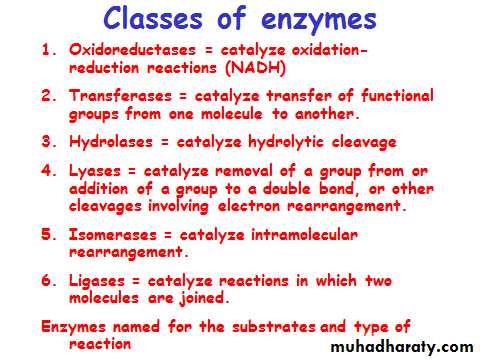
Enzymes
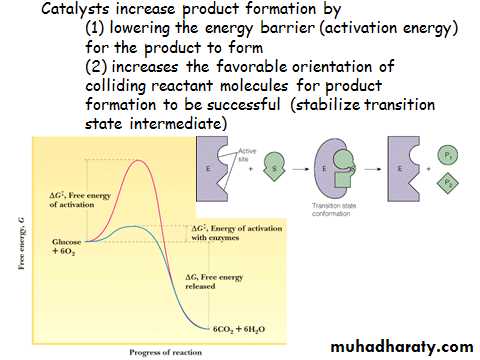
Catalytic Power
Enzymes can accelerate reactions as much as 1016overuncatalyzedrates!•Urease is a good example:
–Catalyzed rate: 3x104/sec
–Uncatalyzed rate: 3x10 -10/sec
–Ratio is 1x1014!
Specificity
•Enzymes selectively recognize proper substrates over other molecules •Enzymes produce products in very high yields -often much greater than 95% •Specificity is controlled by structure -the unique fit of substrate with enzyme controls the selectivity for substrate and the product yield
Co-enzymes
•Non-protein molecules that help enzymes function•Associate with active site of enzyme
•Enzyme + Co-enzyme = holoenzyme
•Enzyme alone = apoenzyme
•Organic co-enzymes –thiamin, riboflavin, niacin, biotin
•Inorganic co-enzymes –Mg ++, Fe++, Zn++, Mn++
Kinetics
•study of reaction rate
•determines number of steps involved
•determines mechanism of reaction
•identifies “rate-limiting” step
Cofactors
Cofactors are organic or inorganic molecules that are required for the activity of a certain conjugated enzymes
• Apoenzyme = enzyme (-) cofactor
• Holoenzyme = enzyme (+) cofactor
• Inorganic cofactors – essential ions
• Organic cofactors – coenzymes
Metabolite coenzymes – synthesized from common metabolites
• Nucleoside triphosphates – (ATP) can donate phosphates, pyrophosphates, adenosyl grroups
• S-adenosylmethionine (SAM) – donates methyl groups
• Nucleotide sugars (uridine diphosphate glucose =
UDP-glucose) - transfer sugars in carbohydrate metabolism