6
Lec.2 DNA REPAIR
Despite the proofreading system employed during DNA
synthesis, incorrect mismatch of base-pairing or insertion of
one or few extra nucleotides occurs. In addition, DNA is
constantly being subjected to environmental insults that cause
the alteration or removal of nucleotide bases. The damaging
agents can be either chemical, for example, nitrous acid, or
radiation, for example, ultraviolet light, which can fuse two
adjacent pyrimidine to each other in the DNA or it could be
high-energy radiation, which can cause double-strand breaks.
Bases are also altered or lost spontaneously from DNA.
if the damage is not repaired, a permanent mutation may be
introduced that can result in a number of deleterious effects,
including loss of control over the proliferation of the mutated
cell, leading to cancer. Luckily, cells are efficient at repairing
the mismatches and other damage done to their DNA.
The repair enzymes are involved in
recognizing the lesion,
excising the damaged section of the DNA strand, &
using the sister strand as a template to fill the gap left by
the excision of the abnormal DNA.
A. Methyl-directed mismatch repair
system
Sometimes replication errors escape the proofreading function
during DNA synthesis, causing a mismatch of one to several
bases this occurs by:
1. Identification of the error (the mismatched strand):
we have a protein called (Mut) protein that identify the
mispaired nucleotides , Mut protein discriminate between the
correct strand and the strand with mismatch, discrimination is
based on the degree of methylation of the two strand (the
newly synthesized and the parentral strand) because the
parental strand is methylated, whereas the newly synthesized
strand is not yet methylated , the parentral strand is assumed to
be the correct while the daughter strand need to be corrected.
Then the next step is:
2. Repair of damaged DNA: When the new strand containing
the mismatch is identified, an endonuclease removes the
mismatched base. The gap left by removal of the mismatched
nucleotide is filled, using the sister
strand as a template, by a
using the enzyme DNA polymerase then the newly
synthesized piece is joined to the original DNA strand by DNA
ligase, A defect in mismatch repair in humans has been shown
to cause hereditary nonpolyposis colon cancer (HNPCC),
one of the most common inherited cancers.
B. Repair of damage caused by ultraviolet
light (UV):
Exposure of a cell to ultraviolet light can result in the covalent
joining of two adjacent (usually thymines), producing a
(dimer) these thymine dimers prevent DNA polymerase from
replicating the DNA strand beyond the site of dimer
formation. Thymine dimers must be excised and the process is
called (nucleotide excision repair) which involves the
following mechanism
Recognition and excision of dimers by UV-specific
endonuclease: First, a UV-specific endonuclease (called
uvrABC ase) recognizes the dimer, and cleaves the damaged
strand at phosphodiester bonds on both side of the dimer. The
damaged oligonucleotide is released, leaving a gap in the
DNA strand,the gap left is filled, by a using the enzyme DNA
polymerase then the newly synthesized piece is joined to the
original DNA strand by DNA ligase.
Ultraviolet radiation and cancer: Pyrimidine dimers can be
formed in the skin cells when exposed to unfiltered sunlight.
In the genetic disease xeroderma pigmentosum, the cells
cannot repair the damaged DNA, resulting in extensive
accumulation of mutations and, consequently, skin cancers.
C. Correction of base alterations (base
excision repair)
The bases of DNA can be altered, either spontaneously or by
the action of chemicals as nitrous acid, as is the case with
cytosine, which slowly undergoes deamination to form uracil
(Normally DNA doesn’t contain uracil), also Bases can also
be lost spontaneously. Lesions involving base alterations or
loss can be corrected by the following mechanisms.
1. Removal of abnormal bases: Abnormal bases, such as
uracil, which can occur in DNA by for example deamination
of cytosine synthesis, are recognized by specific glycosylases
that cleave them from the strand. This leaves an apyrimidinic
or apurinic site, called an AP-site.
2. Recognition and repair of an AP-site: Specific enzyme
called AP- endonucleases recognize that a base is missing and
initiate the process of excision and gap filling A deoxyribose-
phosphate lyase removes the single, empty, sugar-phosphate
residue. DNA polymerase and DNA ligase complete the
repair process.
D .Repair of double-strand breaks
High-energy radiation or oxidative free radicals can cause
double-strand breaks in DNA, which are potentially lethal to
the cell. Double-strand breaks also occur naturally during gene
rearrangements. Double-strand DNA breaks cannot be
corrected by the previously described strategy of excising the
damage on one strandand using the remaining strand as a

7
template for replacing the missing nucleotides. Instead,
double-strand breaks are repaired by one of two systems.
The first is nonhomologous end-joining repair, in which the
ends of two DNA fragments are brought together by a group
of proteins. This system does not require that the two DNA
sequences have any sequence homology. However, this
mechanism of repair, which is the main repair mechanism in
humans, is error prone and mutagenic. Defects in this repair
system are associated with a predisposition to cancer and
immunodeficiency syndromes.
The second repair system is homologous recombination
repair, uses the enzymes that normally perform genetic
recombination between homologous chromosomes during
meiosis,this system is less error prone because any DNA that
was lost is replaced using DNA as template
Transcription
Transcription (RNA synthesis):
Is the process of RNA synthesis directed by a DNA ,DNA is
copied to RNA by an enzyme called RNA polymerase and
occurs in three phases: initiation, elongation and termination.
RNA structures: The DNA and RNA polynucleotide chains
have similar structures except for the presence of uridine in
RNA (instead of thymine in DNA) and for the presence of the
ribose sugar in the RNA instead of deoxyribose in the DNA,
also DNA present as double strand while RNA as single
strand.
The major types of RNA are
rRNA is a component of the ribosomes.
tRNA serves as an molecule that carries a specific amino
acid to the site of protein synthesis.
mRNA carries genetic information from the DNA to the
cytosol, where it is used as the template for protein
synthesis.
small nucleus(snRNA) &small nucleolus(snoRNA)
During transcription, a DNA sequence is read by an RNA
polymerase, which produces a complementary, antiparallel
RNA strand. transcription results in an RNA complement that
contains uracil (U) instead of thymine (T) .
There are four distinct classes of RNA polymerase in the
eukaryotic cells.
1) RNA polymerase I: This enz synthesizes the rRNA.
2) RNA polymerase II: This enzyme synthesizes the mRNAs
that are subsequently translated to produce proteins.
Polymerase II also synthesizes certain small nuclear RNAs
(snRNA)
3) RNA polymerase III: This enzyme produces tRNAs, and
some snRNAs.
4) 4.Mitochondrial RNA polymerase
Transcription is the first step in gene expression,The part of
DNA transcribed into an RNA molecule is called a
transcription unit and encodes at least one gene. If the gene
transcribed encodes a protein, the result of transcription is
messenger RNA (mRNA), which will then be used to create
that protein by the process of translation. Alternatively, the
transcribed gene may encode for ribosomal RNA (rRNA) or
transfer RNA (tRNA)
During DNA replication the whole double stranded genome of
DNA needs to be replicated, but during transcription only
small portion of genome is transcribed in response to need
.
A transcription unit encoding for a protein contains :
The coding sequence: which is the sequence that will later
translate into the protein.
The regulatory sequences that regulate the synthesis of that
protein. The regulatory sequence before (upstream from)
the coding sequence is called the five prime untranslated
region (5'UTR), and the sequence following (downstream
from) the coding sequence is called the three prime
untranslated region (3'UTR)..
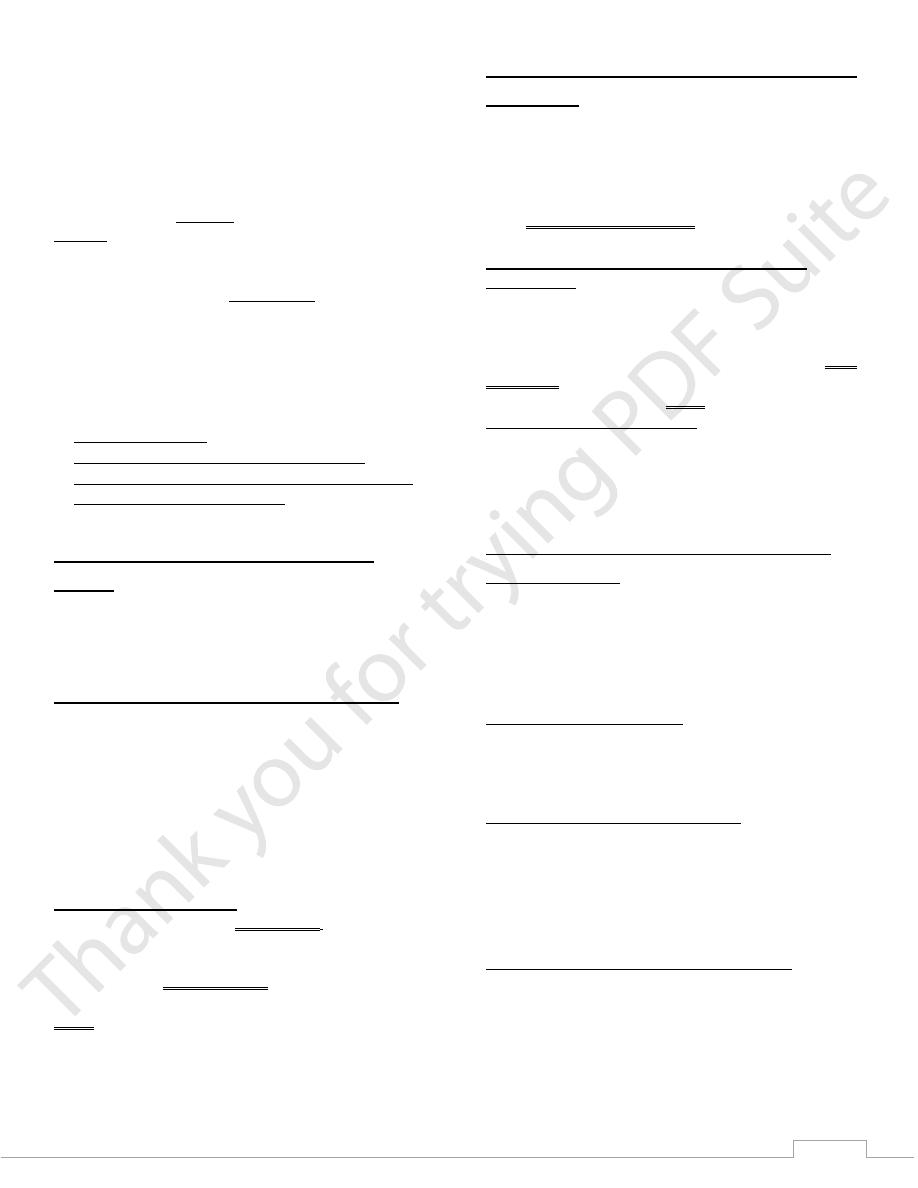
Although DNA is arranged as two antiparallel strands in a
double helix, both can be used in transcription but only one of
the two DNA strands is used for transcription (called the
template strand or the antisense strand), this is because RNA is
only single-stranded, the other DNA strand is called the(
coding strand or the sense strand)
Transcription is divided into: initiation, elongation and
termination
Initiation:
The initiation of transcription, requires the presence of a
1. Promoter Sequence
2. Transcription Factors
3. RNA Polymerase
4. Activators and Repressors.
Promoters are special nucleotides sequence in DNA that
allows transcription Promoters are usually rich in thymine and
adenine in repeating pattern and called TATA box, another
consensus sequence or promoter is known as the CAAT box,
in other genes no TATA box is found instead a GC-rich region
(GC box) may be found as promoters ,promoters are located
immediately upstream of transcription start site, which is the
site where the first ribonucleotide unit is paired with the
template strand .
RNA polymerase cannot recognize the promoter sequences.
Instead, a special collection of proteins called transcription
factors (TF) mediate the binding of RNA polymerase and the
initiation of transcription, these transcription factors
recognizes the promoter & bring RNA Polymerase II to the
8
promoter, also TF regulate the frequency of initiation, and
mediate the response to signals as hormones.
Only after certain transcription factors are attached to the
promoter then the RNA polymerase bind to it. The completed
assembly of transcription factors and RNA polymerase bind to
the promoter, forming a transcription initiation complex
The TATA box, as a promoter, is the binding site for a protein
known as TATA-binding protein (TBP) which is a type of TF
then more transcription factors(TFIIB, TFIID, TFIIE, TFIIF,
TFIIH) (TF means transcription factor) and RNA polymerase
combine around the TATA box in a series of stages
to form a
preinitiation complex
. Some transcription factor, has a
helicase activity and so is involved in the separating of
double-stranded DNA,
Specific transcription factors bind to other proteins
(Coactivators) recruit them to the transcription complex.
Simple diagram of transcription initiation. RNAP = RNA
polymerase
Transcription
RNA polymerase
closed promoter complex
open promoter complex
initiation
elongation
termination
RNA product
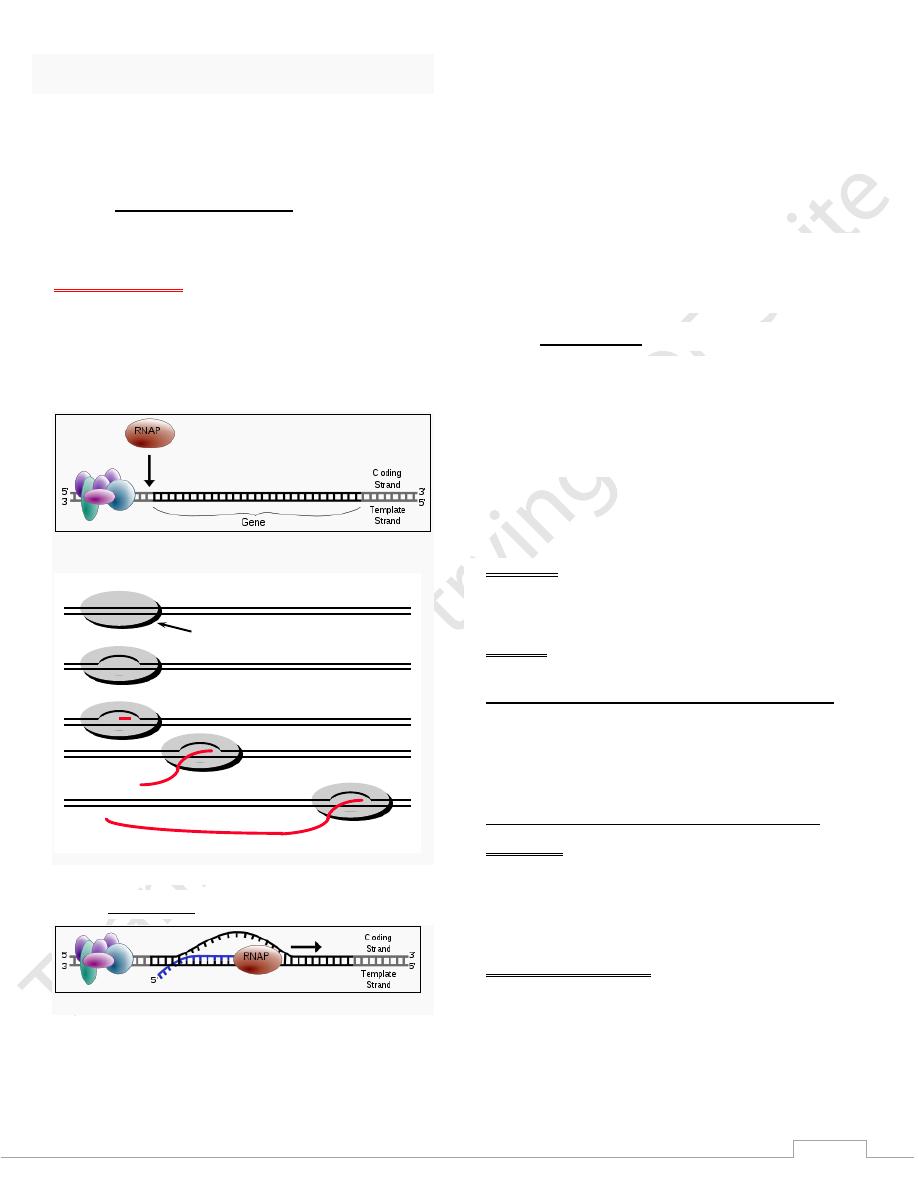
Elongation
Simple diagram of transcription elongation
Once the promoter region has been recognized, the RNA
polymerase begins to synthesize a transcript of the DNA
sequence. Unlike DNA polymerase, RNA polymerase does not
require a primer and has no ability to repair mistakes in the
RNA, as does DNA polymerase during DNA synthesis .One
strand of the DNA, the template strand (or noncoding strand),
is used as a template for RNA synthesis. As transcription
proceeds RNA polymerase traverses the template strand and
uses base pairing complementarily with the DNA template to
create an RNA copy. This produces an RNA molecule from 5'
→ 3', an exact copy of the coding strand (except that thymines
are replaced with uracils, and the nucleotides are composed of
a ribose (5-carbon) sugar where DNA has deoxyribose .
mRNA transcription can involve multiple RNA polymerases
on a single DNA template and multiple rounds of transcription
so many mRNA molecules can be rapidly produced from a
single copy of a gene.
Termination
Transcription termination involves cleavage of the new
transcript followed by addition of number of(Adenine) at its
new 3' end, in a process called polyadenylation.
Transcriptional control of genetic expression is vital for
cellular functions, and many diseases and cancers are results
of defects in the transcriptional control of essentials genes
Termination provides a point at which transcription can be
controlled. Many factors act by decreasing (anti-termination)
or enhancing termination rates at certain RNA sequences,
thereby controlling the expression rates of genes.
Enhancers are special DNA sequences that increase the
rate of initiation of transcription by RNA polymerase II.
Enhancers on the same chromosome as the gene whose
transcription they stimulate (cis-acting).
Silencers reduce the level of gene expression.
Posttranscriptional modification of RNA
A primary transcript is a linear copy of a transcriptional
segment of DNA between specific initiation and termination
sequences. The primary transcripts of RNAs are
posttranscripitinally modified.
A. Post transcriptional modification of messenger RNA
1) "Capping": This process is the first of the processing
reactions for mRNA ,The cap is added by the enzyme
guanylytransferase the addition of this7- methylguanosine
cap
permits the initiation of translation and
help stabilization of the mRNA if the mRNA lack the cap
it is not efficiently translated.
2) Addition of a poly A- tail: Most mRNA have a chain of 40 to
200 adenine nucleotides attached to the 3'-end This poly-A tail
is not transcribed from the DNA, but rather is added after
transcription. These tails help stabilize the mRNAs and
facilitate their exit from the nucleus and aid translation, After
the mRNA enters the cytosol, the poly-A tail is gradually
shortened.

9
3) Removal of introns: Modification involves the removal of
introns from the primary transcript. (Introns are RNA
sequences, which do not code for protein) The remaining
coding sequences, called the exons, are joined together to
form the mature mRNA. The process of removing introns and
joining exons is called splicing The molecular machine that
accomplishes these tasks is known as the spliceosome.This
process make the mRNA functional and ready for
translation.
a.Role of small nuclear RNAs (snRNAs): snRNAs, in
association with proteins, form particles (snRNP). These
facilitate the splicing(joining) of exon segments by forming
base pairs with the consensus sequences at each end of the
intron .
Effect of splice site mutations: Mutations at splice sites can
lead to improper splicing and the production of aberrant
proteins. It is estimated that fifteen percent of all genetic
diseases are a result of mutations that affect RNA splicing.
4) Alternative splicing of mRNA molecules: The pre- mRNA
molecules from some genes can be spliced in two or more
alternative ways in different tissues. This produces multiple
variations of mRNA and therefore, of its protein product, this
appears to be a mechanism for producing a diverse set of
proteins from a limited set of genes. For example, different
types of muscle cells all produce the same primary transcript
from the tropomyosin gene. However, different patterns of
splicing in the different cell types produce a family of tissue-
specific tropomyosin protein molecules.
B. Post transcriptional modification of Ribosomal RNA
rRNA are synthesized from long precursor molecules called
pre-rRNA. The pre-rRNAs are cleaved by ribonucleases to
yield intermediate-sized pieces of rRNA, which are further
processed "trimmed by exonucleases and modified at some
base and sugar facilitated by small nucleolar RNA to produce
the required rRNA.
C. Post transcriptional modification of Transfer RNA
tRNAs are also made from longer precursor molecules that
must be modified . An intron must be removed from the
anticodon by nucleases, some sequences at both the 5'- and the
3'-ends of the molecule must be removed.
