83
Composition of DNA:
DNA, the genetic material, is found principally in the chromosomes, which are located in the
nucleus of a cell (Small amounts of DNA are also found in mitochondria).
It has three basic components:
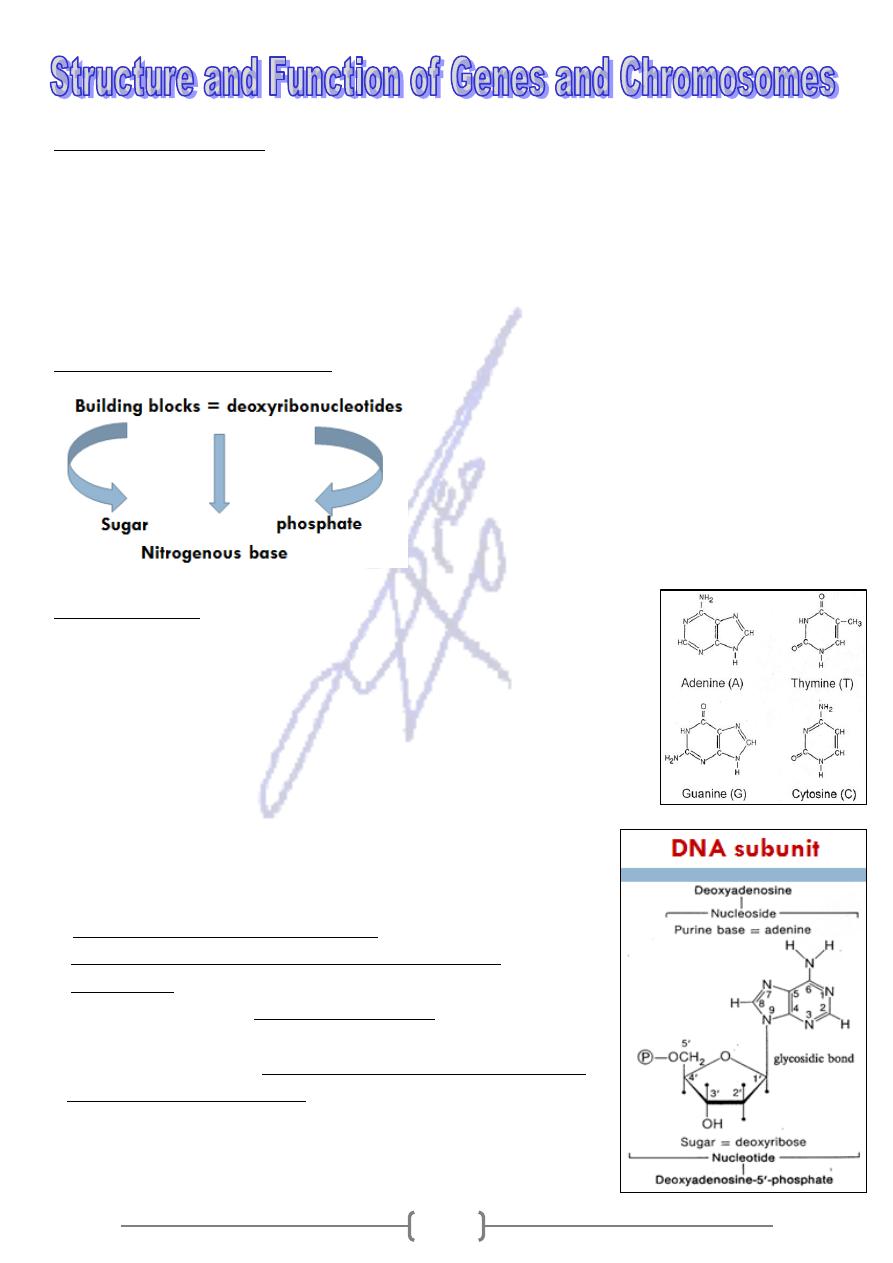
1- the pentose sugar (deoxyribose)
2- a phosphate group, and
3- four types of nitrogenous bases.
DNA (deoxyribonucleic acid)
Nitrogen Bases
*
Two of the nitrogen bases, cytosine and thymine, have single
heterocyclic ring of carbon and nitrogen atoms called pyrimidines.
* The other two bases, adenine and guanine, have double carbon-
nitrogen rings purines.
* The four bases are commonly represented by their first letters: C, T, A,
and G.
* The sugar in DNA molecule is deoxyribose, a 5 carbon sugar, and
successive sugar residues are linked by covalent phosphodiester
bonds. Covalently attached to carbon atom number 1' of each
sugar residue is a nitrogen base.
nucleoside
nucleoside. A
a
A sugar with an attached base is called
*
with a phosphate group attached at carbon atom 5' or
A strand.
the basic repeat unit of DN
a nucleotide,
3'constitutes
on the 2' carbon,
lacks a hydroxyl group
deoxyribose sugar
* The
hence deoxy.
that position in
presence of a hydroxyl at
* This is in contrast to the
The composition of RNA molecules
.
RNA
sugar found in
ribose
the
is similar to that of DNA molecules, but differs in that they contain
uracil (U) instead of thymine.
84
DNA structure:
alternating sugar
of a DNA molecule and of a RNA molecule consists of
linear backbone
The
*
In each case the bond linking an individual sugar residue to the
residues and phosphate groups.
neighboring sugar residue is a 5' - 3' phosphodiester bond. This means that a phosphate group
links carbon atom 3' of a sugar to carbon atom 5' of the neighboring sugar. So the polynucleotide
chain is formed by linking nucleotides through 5‘, 3' -phosphodiester bonds.
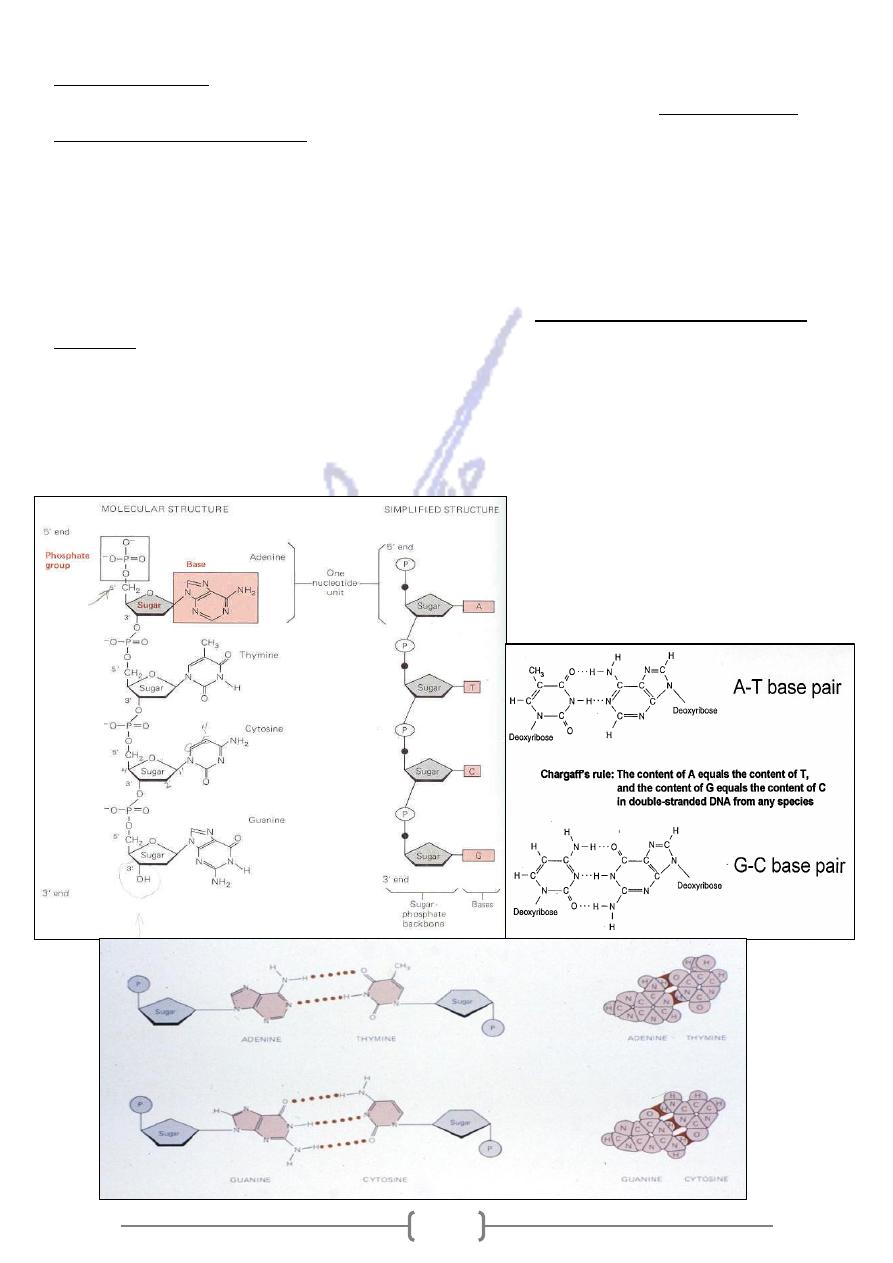
* The structure of DNA is double helix in which two DNA molecules (DNA strand) are held
together by weak hydrogen bonds to form a DNA duplex. Hydrogen bonding occurs between
-
DNA duplex according to Watson
aterally opposed bases, base pairs of the two strands of the
l
bonds.
-
T) base pair share two H
-
Thymine (A
-
Adenine
:
Crick rules
Guanine-Cytosine (G-C) base pair share three H- bonds.
* G:C- rich regions of DNA are more stable.
* Because of base pairing, the polynucleotide chains in double-stranded DNA are
complementary to each other.
85
* As a result, the base composition of DNA from different cellular sources is not random:
the amount of adenine equals that of thymine, and the amount of cytosine equals that of
guanine, [A] = [T] and [G] = [C] [purines] = [pyrimidines]. For example, if a source of DNA is
quoted as being 42% GC, the base composition can be inferred to be: G 21%,C 21%, A 29%,
T 29%.
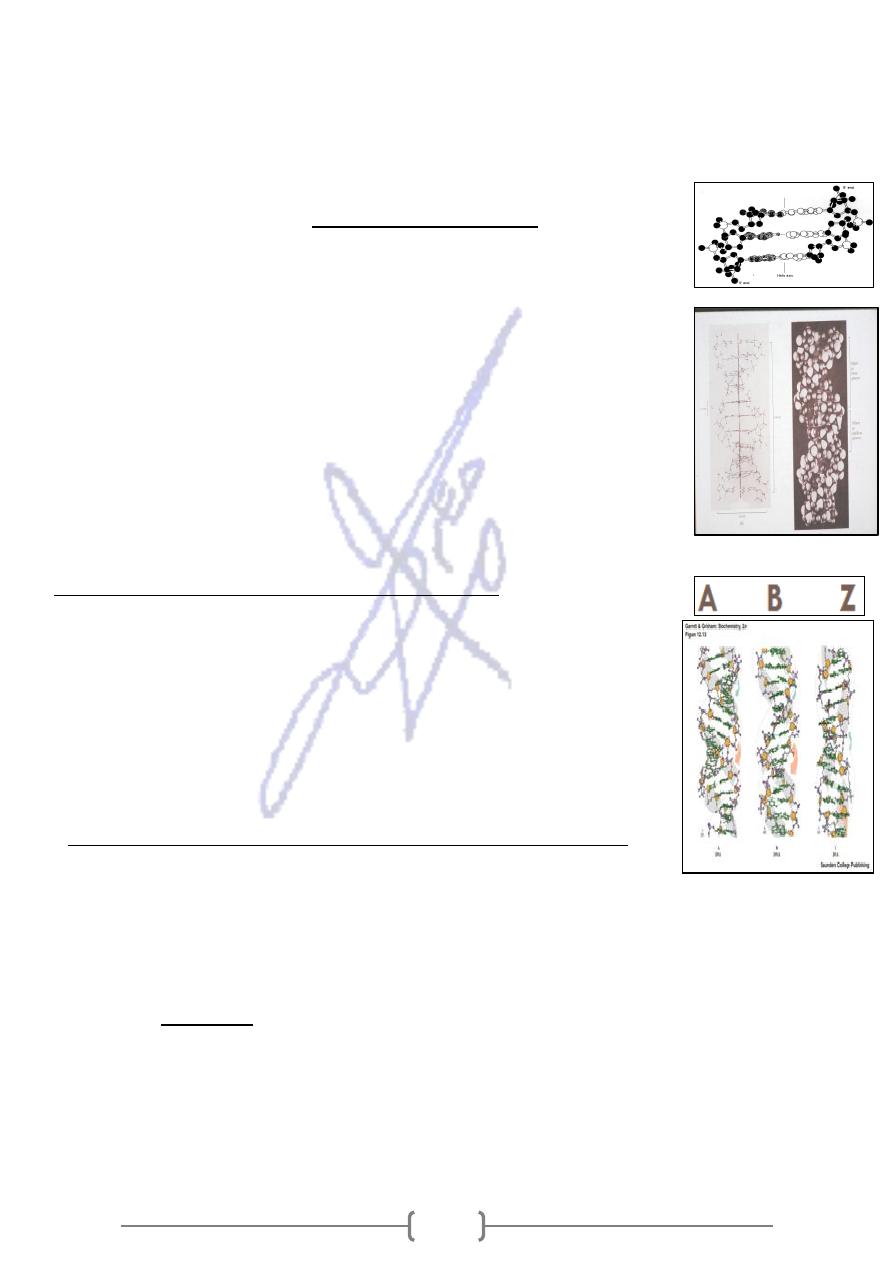
double helix.
three base pairs in the DNA
* This slide shows a side view of
Note the base-pair stacking interactions, the hydrophobic interior, and the
phosphates on the exterior.
* The DNA double helix can be envisioned as a twisted ladder, the bases
make up the rungs of the ladder.
* The two sides of the ladder are composed of the sugar projecting from each
side of the ladder, at regular intervals.
* The nitrogen base projecting from one side is bound to the base projecting
from other side by relatively weak hydrogen bonds, the phosphate
components held together by strong phosphodiester bonds
DNA has different types of helical structure
* A-DNA and B- DNA are both right- handed helices (ones in which the helix
spirals in a clockwise direction as it moves away from the observer). They
have respectively 11 and 10 base pairs per turn.
* Z- DNA is a left- handed helix which has 12 base pairs per turn.
* Under physiological conditions, most of the DNA in a bacterial or
eukaryotic genome is of the B- DNA form in which each helical strand
has a pitch (the distance occupied by a single turn of the helix of 3.4 nm).
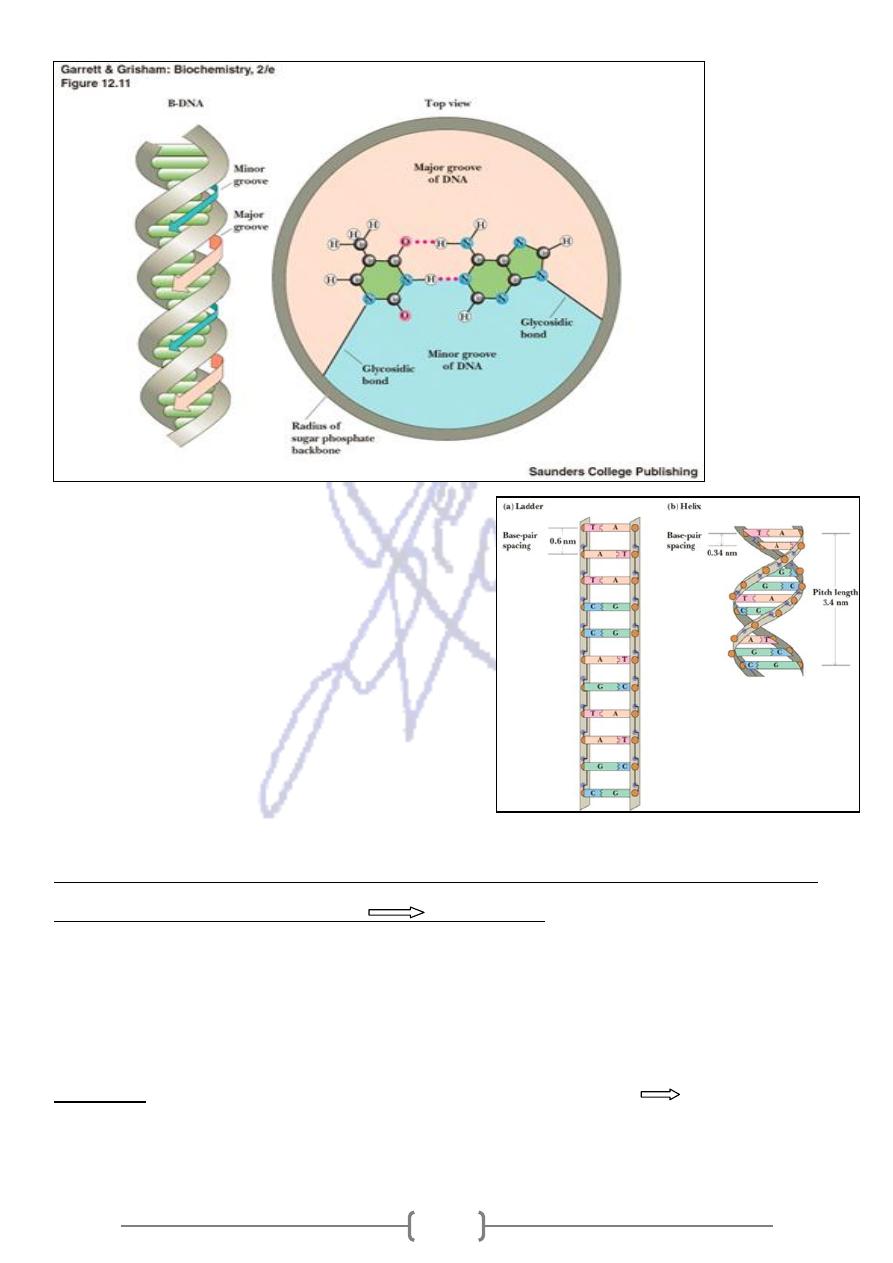
A major groove and a minor
has two kinds of grooves in a helix.
DNA
-
B
*
groove, and has 10 base pairs per one turn of the double helix.
* The "tops" of the bases line the "floor" of the major groove may provide:
- a binding sites, so that the regulatory proteins (transcription factors) can recognize the
pattern of bases - and also H-bonding possibilities in the major groove
* DNA that is overwind or underwind, with fewer than or more than 10 base pairs per turn, is
.
supercoiled
"
said to be
86
Why a helix? Why not a ladder?
*
The heterocyclic bases have flat surfaces which
are hydrophobic
* To exclude water from between the rings, we
should bring the bases closer together
* One way to model them closer together is to
“twist” the ladder into a helix. The base-pair
spacing in Ladder is 0.6 nm, while in case of
Helix, the base-pair spacing is 0.34 nm.
escribe a DNA sequence by writing the sequence of bases of
It is usual to d
one strand only, and in the 5' 3' direction
* As the phosphodiester bonds link carbon atoms number 3' and number 5' of successive sugar
residues, one end of each DNA strand, the so- called 5‘ end, will have a terminal sugar residue
in which carbon atom number 5' is not linked to a neighboring sugar residue. The other end is
defined as the 3' end because of a similar absence of phosphodiester bonding at carbon atom
number 3' of the terminal sugar residue. The two strands of a DNA duplex are said to be
because they always associate (anneal) in such way that the 5' 3' direction of
antiparallel
one DNA strand is the opposite to that of its partner.
* This is the direction of synthesis of new DNA molecules during DNA replication, and also of
the nascent RNA strand production during transcription.
87
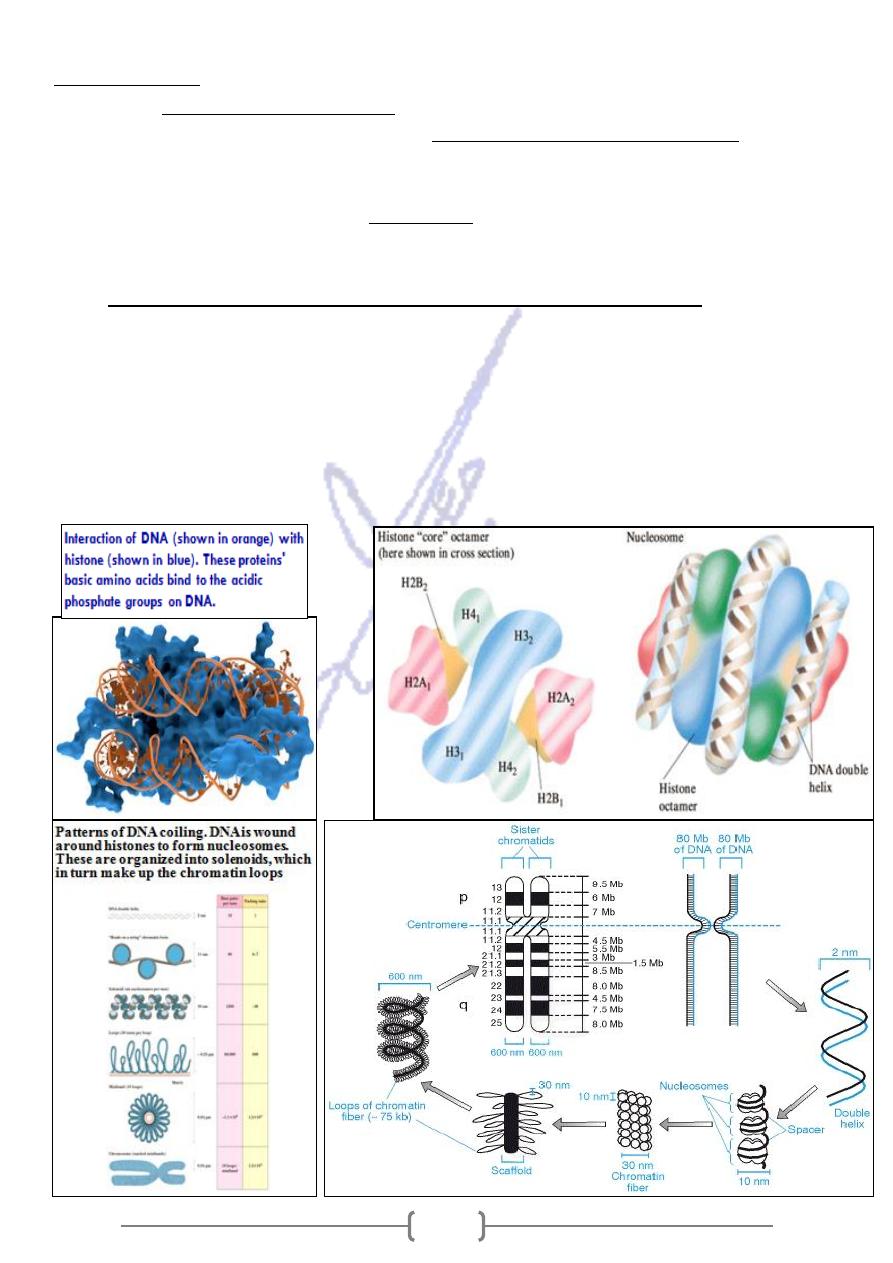
DNA Packaging
. To package all of this DNA into a tiny cell nucleus, it
DNA total length is ~2 meters
* The human
. This
nucleus that is ~ 5 microns in diameter
is coiled at several levels. It is packaged into a
represents a compression of more than 100,000 fold.
* DNA is packed inside the nucleus in association with a number of proteins, which are
. Each nucleosome is made up a histone.
nucleosomes
extensively coiled and folded forming
* Histone octamer mainly made up of histones H2A, H2B, H3 and H4. The DNA is wound
around a histone protein. About 140-150 DNA bases are wound around each histone core and
next 50 bp form a spacer element to links one nucleosome to another.
e
th
then
* Also interacting with another histone (H1) forming a thicker fibre consisting of six
nucleosomes, known as the solenoid.
* The solenoids themselves are organized into chromatin loops, which are attached to a
protein scaffold (one of these proteins is the enzyme topoisomerase type II).
* Each of these loops contains approximately 100,000 bp or 100 kb of DNA.
* So DNA is a tightly coiled structure. This coiling occurs at several levels:
- the nucleosome, - the solenoid, and - the 100- kb loops.
88
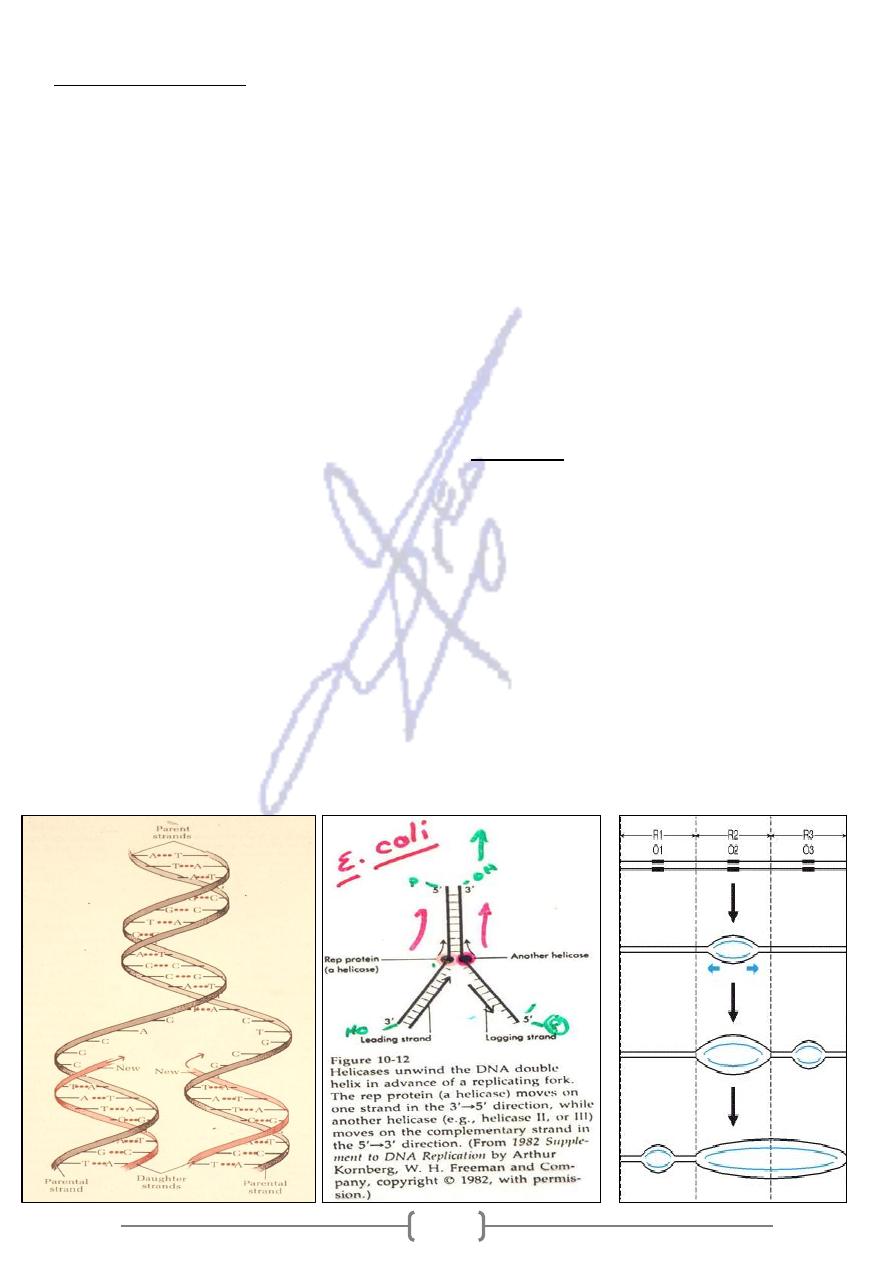
Replication of DNA
*
As cells divide, identical copies of DNA must be made and incorporated into the new cells.
During this process of DNA replication:
- the two DNA strands of each chromosome are unwound by a helicase enzyme
- Each DNA strand directs the synthesis of a complementary DNA strand to generate two daughter
DNA duplexes, each of which is identical to parent molecule.
- Hence, the newly replicated chromosome consists of one old and one new DNA strand.
- Because each strand of DNA serves as a template for the production of a complementary strand,
DNA replication is called semiconservative.
* DNA replication is initiated at specific points called "origins of replication". Starting from
such origin, the initiation of DNA replication results in a replication fork, where the parental
DNA duplex bifurcates into two daughter DNA duplex.
, but act individually as
antiparallel
* As the two strands of the parental DNA duplex are
templates for the synthesis of complementary antiparallel daughter strand
* The rate of DNA replication in humans about 40-50 nucleotides per second, it is slow.
* In bacteria the rate is much higher, reaching 500 to 1,000 nucleotide per second.
* The human chromosomes have as many as 250 million nucleotides, so it is time-consuming
process if it processed linearly, (because in a chromosome of this size, a single round of
replication would take almost two months).
- Instead DNA replication in mammalian chromosomes precedes bidirectionally at many
different points along the chromosome, the resulting multiple separations of the DNA strands
called replication bubbles.
- The distance between adjacent replication origins is about 150 kb.
- The resulting "replication bubbles" then fuse together or merge completing the synthesis of
the daughter chromosomes.
89
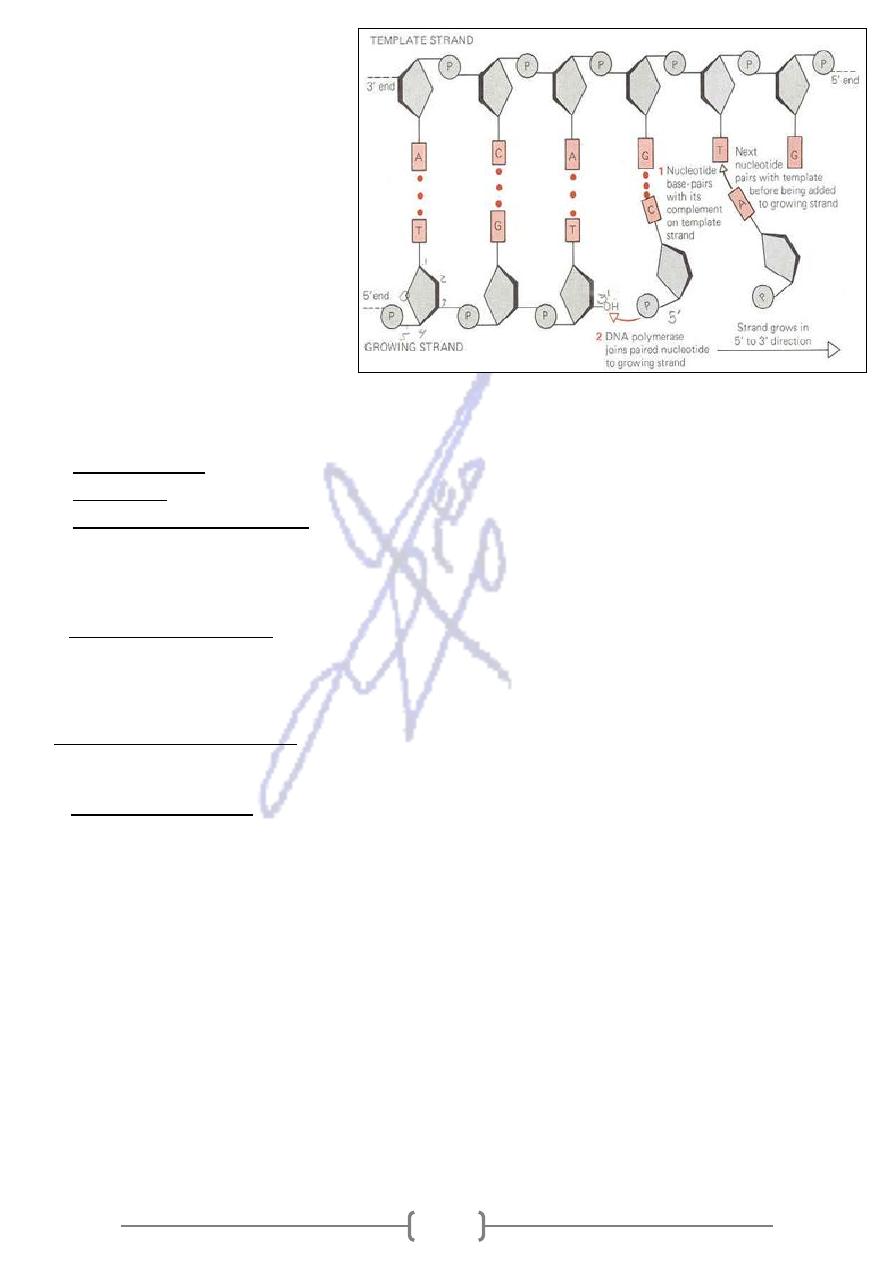
* The direction of chain growth
leading strand synthesis and the
other daughter strand synthesis
(both are in a 5' to 3' direction).
* DNA polymerase is one of the key
replication enzymes. DNA
polymerase moves continuously
along the leading strand.
* All DNA polymerases require a
primer (or a growing DNA chain)
with a free 3' OH.
The requirements of DNA replication:
Helicase enzyme
1.
(or a growing DNA chain) with a free 3' OH.
DNA primer
2.
phosphate,
5’
-
includes: (deoxyadenosin
Triphosphate polynucleotides
3.
deoxythymidin-5’ phosphate, deoxyguanosine-5’ phosphate, deoxycytidine-5’ phosphate).
The triphosphate nucleotides first is cleaved to monophosphate, then added to the end of
the new strand.
DNA polymerase enzyme
4.
DNA-Modifying Enzymes
1. Nucleases: are enzymes that cut DNA strands by catalyzing the hydrolysis of the
, there are two types:
phosphodiester bonds
* Exonucleases: that hydrolyses nucleotides from the ends of DNA strands.
* Endonucleases: cut within strands.
2. DNA ligases: can rejoin cut or broken DNA strands. Ligases are particularly important in
lagging strand in bacterial DNA replication, as they join together the short segments of DNA
produced at the replication fork into a complete copy of the DNA template. They are also used
in DNA repair and genetic recombination.
3. Topoisomerases: are enzymes with both nuclease and ligase activity.
* These enzymes change the amount of supercoiling in DNA.
* Some of these enzymes work by cutting the DNA helix and allowing one section to
rotate, thereby reducing its level of supercoiling; the enzyme then seals the DNA break.
* Topoisomerases are required for many processes involving DNA, such as DNA
replication and transcription.
91
4. Helicases: are proteins that are a type of molecular motor. They use the chemical energy in
nucleoside triphosphates, predominantly ATP, to break hydrogen bonds between bases and
unwind the DNA double helix into single strands.
* These enzymes are essential for most processes where enzymes need to access the DNA
bases.
5. Polymerases: the key replication enzymes ,that synthesize polynucleotide chains from
nucleoside triphosphates.
These enzymes function by:
A. It travels along the single DNA strand (template), adding free nucleotides to the 3' end of the
new strand. Nucleotides can be added only to the 3' end of the strand, so replication always
proceeds from the 5' to the 3' end.
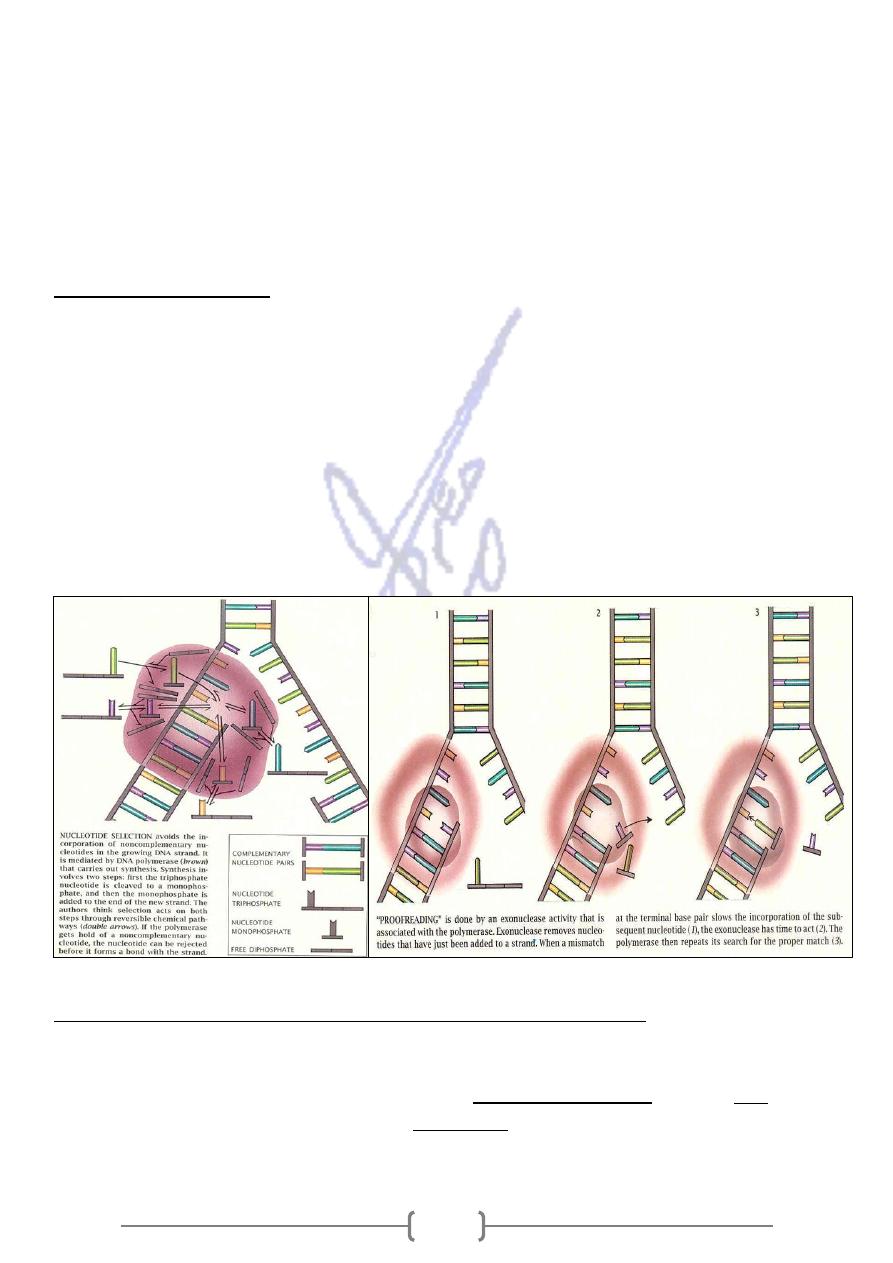
B. In addition to adding new nucleotides, DNA polymerase performs part of a proofreading
activity. Here, the polymerase recognizes the occasional mistakes in the synthesis reaction by
the lack of base pairing between the mismatched nucleotides. If a mismatch is detected, a 3′ to
5′ exonuclease activity is done and the incorrect is repair.
* When a DNA replication error is not successfully repaired, a mutation has occurred causing
genetic diseases.
Polymerases are classified according to the type of template that they use:
* In DNA replication, a DNA-dependent DNA polymerase makes a copy of a DNA sequence.
* RNA-dependent DNA polymerases are a specialized class of polymerases that copy the
viral
which is a
reverse transcriptase
to DNA. They include,
sequence of an RNA strand in
.
retroviruses
enzyme involved in the infection of cells by
* Telomerase: which is required for the replication of telomeres. Telomerase is an
unusual polymerase because it contains its own RNA template as part of its structure.
91
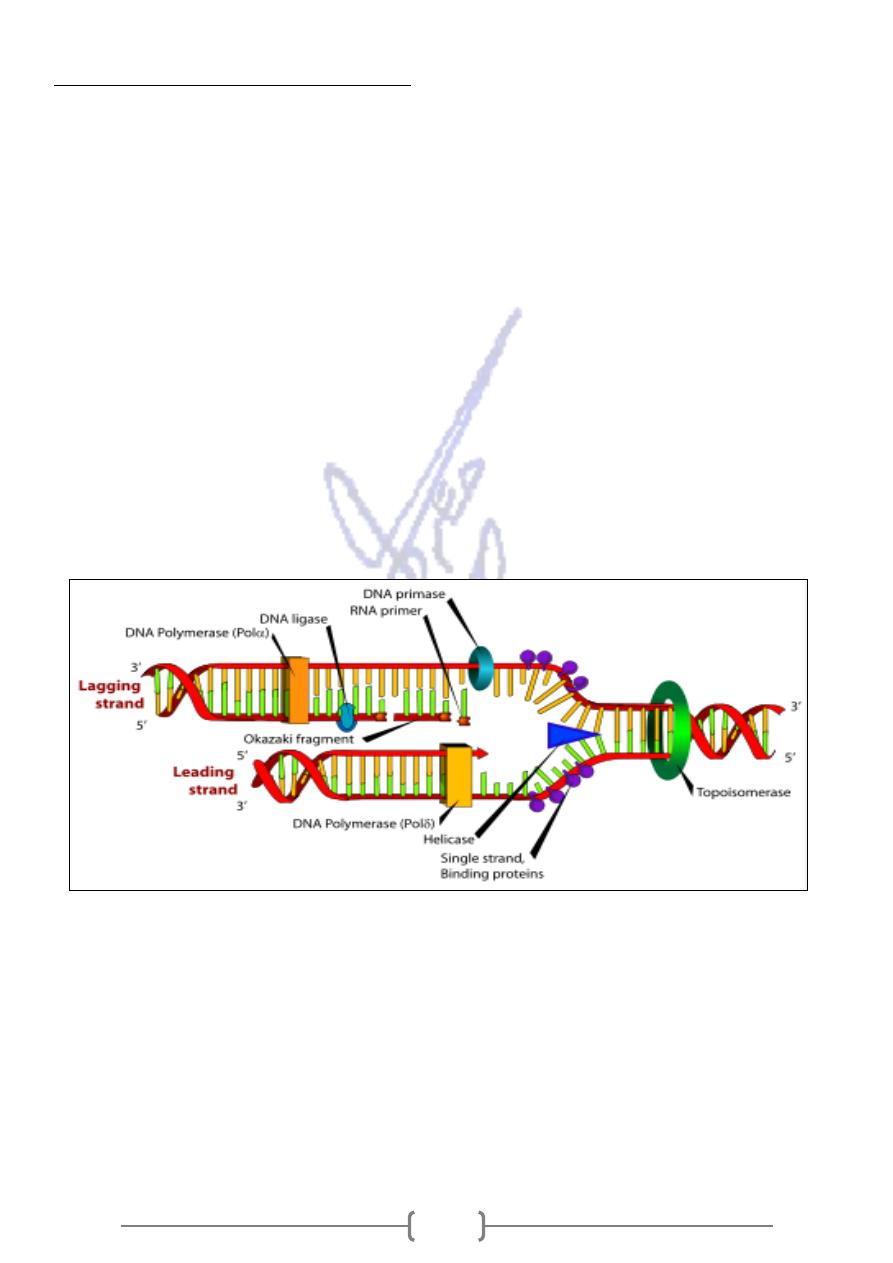
Replication of Bacterial chromosome
* The circular E. coli chromosome has one origin of replication (ori).
- Initiation of replication begins with the binding of proteins to the ori sequence act as coalesce,
the adjacent DNA is forced to undergo melting into single strands.
- This allows the DNA "helicase" or DNA unwinding enzyme to bind the single-stranded DNA
and further unwind the double helix.
- Because DNA synthesis always proceeds in a 5' to 3' direction, and because the two DNA
strands are arranged antiparallel with respect to each other, only one of the two newly
synthesized strands can be made "continuously" as the DNA polymerase moves away from
the origin of replication and more DNA template is exposed. The other strand has to be made
"discontinuously" in short pieces.
* The continuously synthesized strand is called the "leading strand." while the discontinuously
strand called the "lagging strand“.
* As further unwinding occurs, another protein "primase" synthesizes a short RNA primer of
about 5 nucleotides. Because the primase synthesizes an RNA strand, it is an RNA
polymerase.The primer provides a free 3' OH end to initiate DNA synthesis
- Once the RNA primer has been synthesized, DNA polymerase can then bind and begin to
synthesize DNA.
- DNA polymerase catalyzes an attack by the 3' OH on the phosphate of the dGTP, forming a 3',
5'-phosphodiester bond, and releasing two molecules of inorganic phosphate.
- All DNA polymerases require a primer (or a growing DNA chain) with a free 3' OH. The new
strand of DNA grows in a 5' to 3' direction.
* The small pieces of DNA that comprise the lagging strand are called "Okazaki fragments."
They are eventually ligated together forming a continuous DNA strand.
92
* As the replication fork moves further, opening up more DNA, another RNA primer has to be
synthesized. Each RNA primer on the lagging strand then serves as a starting point for the
initiation of DNA synthesis (Okazaki fragment).
* The main DNA polymerases required for DNA replication in E. coli are DNA polymerases I, II
and III.
- Both I and III have 5' to 3' polymerizing activity and 3' to 5' proofreading activity.
- DNA polymerase I also removes the RNA primer and is also a DNA repair enzyme, and
thus require a 3' to 5' exonuclease activity.
at least five DNA
human cells have
DNA polymerases in E.coli,
three
* In contrast to just
(alpha, beta, gamma, delta, epsilon).
polymerases
Plasmid
* Is an extra-chromosomal DNA molecule separated from the chromosomal DNA, which is
capable of replicating independently of the chromosomal DNA.
- In many cases, it is circular and double- stranded.
- Plasmid usually occurs naturally in bacteria, but are sometimes found in eukaryotic organisms.
- Plasmids are considered transferable genetic elements or ”replicons" capable of autonomous
replication within a suitable host.
The major differences between Plasmids and Chromosomes:
* Plasmids have much less base pairs than chromosomes
* Are easily transferred
* Usually contain non- essential genes
* Function can be lost or gained without harming to organism
* Are usually found in lower organisms
