ENZYMES
BY
PROFESSOR
Dr
.
MUZHIM
ALKABBAN

• Enzymes are protein catalysts,
increase the velocity of chemical
reactions. They are not consumed
during the catalytic reactions.
• Nomenclature:
• Two types of names are given for
each enzyme.

• 1. Recommended name which is short.
• 2. Systematic name which is more
complete.
• Recommended name:
Most commonly used, have the suffix [-
ase], attached to the substrate of the
reaction. Ex: Glucosidase; Urease;
Sucrase.
The suffix may attached to the
description of the action performed.

• Ex: Lactate dehydrogenase; adenylate
cyclase.
• Some enzymes retain their original names
which has no hint of the associated
enzyme reaction.
• Ex: Trypsin; pepsin.
• Systematic name:
• The enzymes are divided to six major
classes in this system, each with
numerous subgroups.

• The suffix [-ase] in this system is attached to
the complete description of the chemical
reaction catalyzed. Ex: D-glyceraldehyde 3-
phosphte: NAD oxidoreductase.
•
1. Oxidoreductase:
Catalyze oxidation-reduction reaction. These
are enzymes that catalyze reactions in which one
substrate is oxidized, acting as a proton donor,
and another substrate is reduced, acting as a
proton acceptor.

• Ex: Dehydrogenase, which convert single
bond to double bond.
• Oxidase, which use O
2
as oxidant.
• Peroxidase, which use H
2
O
2
as oxidant.
• Hydroxylase, which introduce hydroxyl
groups.
• Oxygenases, which introduce molecular
oxygen in place of a double bound in the
substrate.

• Lctate + NAD
+
←--------→ Pyruvate + NADH + H
+
CH
3
-CH
-
OH-COO
-
+ NAD
+
↔ CH
3
-C
=
O-COO
-
+NADH+H
+
•
2. Transferases:
Enzymes that catalyze transfer of C, N or P
containing groups. Ex: Transfer of one carbon group
(methyl group), aldehydic or ketotic groups or
phosphoryl groups from one substrate to another.
UDP-galactose + glucose
←----------------→ UDP +
galactosylglucose (lactose)
Enzyme: UDP galactose-glucose galactosyl
transferase enzyme.

• Serine + Tetrahydrofolate
←------→ Glycine + Methyl
tetrahydrofolate
CH2-OH-CH-NH3+-COO
-
+THF
↔CH
2
-NH3+-COO
-
+
THFCH
2
• Enzyme: serine hydroxymethyl transferase.
•
3. Hydrolases:
• Enzymes that catalyze cleavage of bonds by
addition of water as C-O, C-N, P-O, C-C and other
single bonds.
• Ex: Esterase: R-C=O-R + H
2
O
←---→ R-C=O-OH + R-
OH
• Glycosidases and phosphatases:
• Urea + H
2
O -------
urease
--------
→ CO
2
+ 2NH
3
• NH
2
-C=O-NH
2
+ H
2
O ----
urease
----
→ CO
2
+ 2NH
3

•
4. Lyases:
• Enzymes that catalyze cleavage bonds as C-C C-S
and certain C-N bonds without the addition of
water, but by elimination reactions to form double
bonds or rings.
• Ex: Decarboxylases as aldolase:
• Fructose 1,6-diphosphate
←------
Aldolase
----
→
Dihydroxyacetonephosphate + glceraldehyde-3-
phosphate.
Pyruvate-
-pyruvate
decarboxylase
→Acetaldehyde+CO
2
CH
3
-C=O-COO
-
---
pyruvate decarboxylase
-
→CH
3
-
C=O
-
H+
CO
2

•
5. Isomerases:
• Enzymes that catalyze the rearrangement of the
atoms of a molecule (catalyze racemization of
geometric isomers).
• Dihydroxyacetone phosphate
←
triose phosphate
isomerase
→ Glyceraldehyde 3-phosphate.
•
6. Ligases:
• Enymes that catalyze the joining of two
compounds so, catalyze formation of bonds
between carbon and O,S,N coupled to hydrolysis of
high-energy phosphatases.

• Also called or known as synthetase. They
couple the hydrolysis of a pyrophosphate in
ATP or other nucleoside triphosphate to a
second reaction in which two molecules are
joined.
• ATP + acetate + CoA ---
→ AMP + Ppi + acetyl
CoA
Enzyme: acetate CoA Ligase
Pyruvate + CO
2
+ ATP---
pyruvate carboxylase-
-
-
→ Oxaloacetate + ADP + Pi
CH
3
-C=O-COO
-
+ CO
2
+ ATP--
pyruvate
carboxylase-
-
→ HOOC-CH
2
-C
=O
-COO
-
+ ADP +
Pi

•
Properties of enzymes:
•
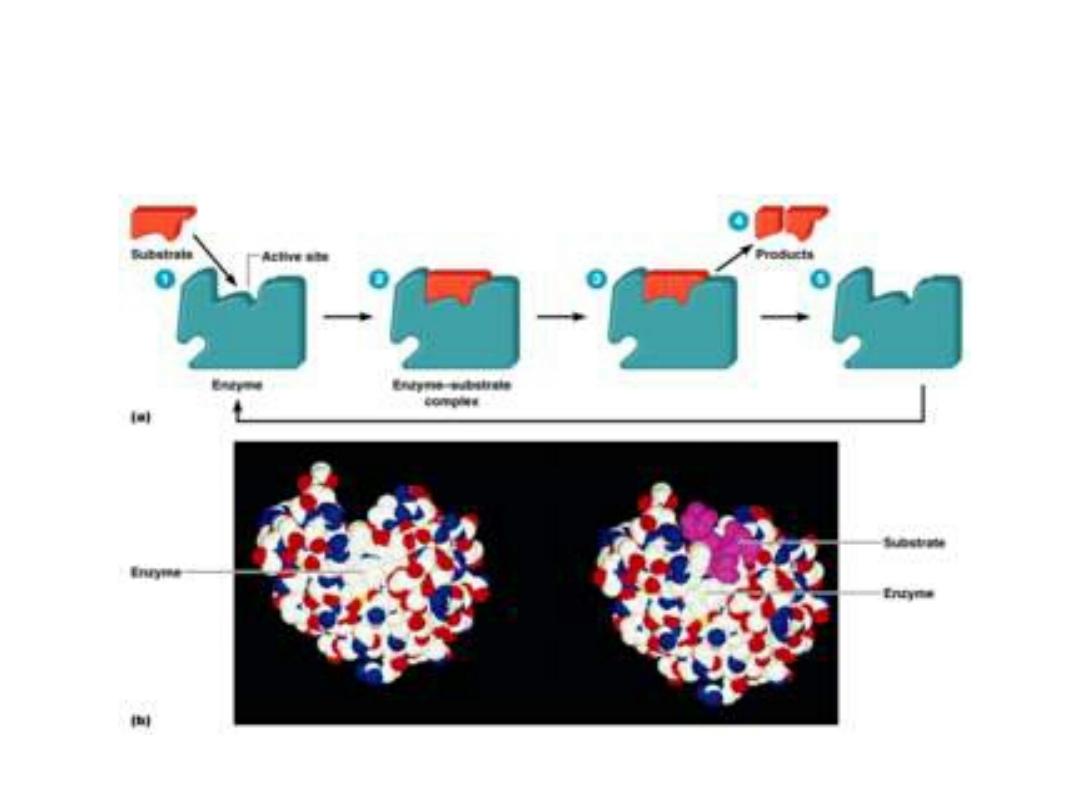
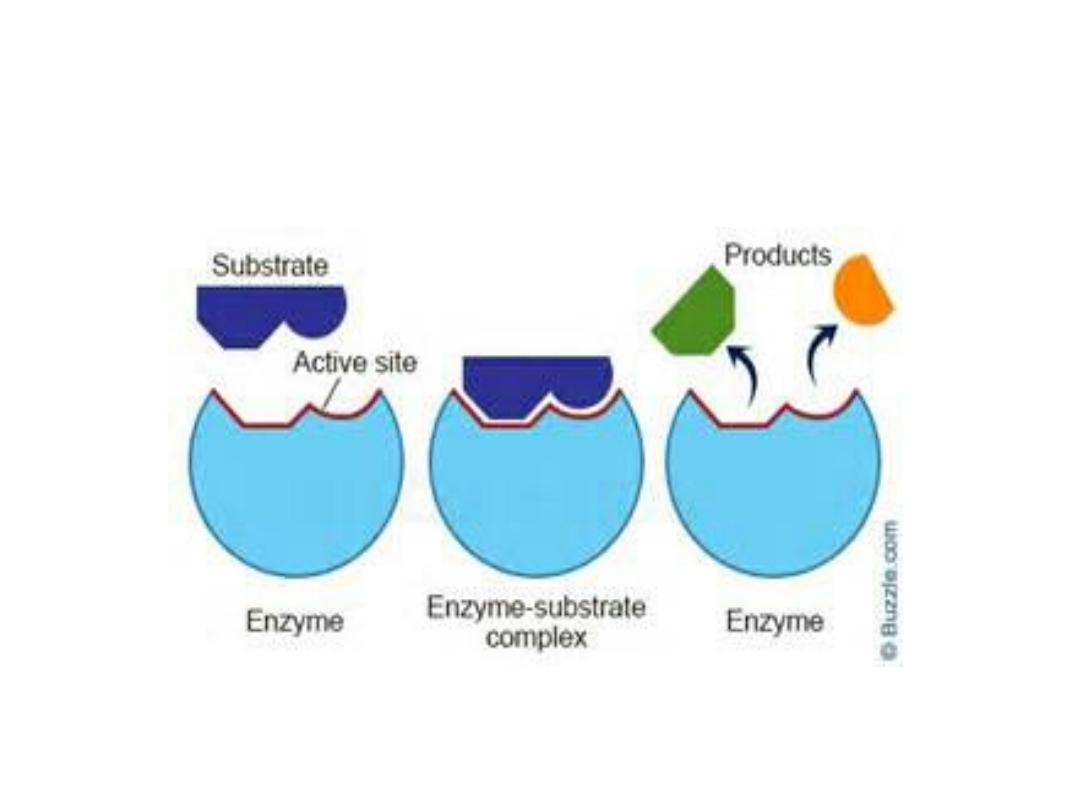
1. Active sites:
• Active site is a special pocket or cleft present on
enzyme molecules. These active sites contain
amino acid side chains which create a three-
dimensional surface complementary site binds to
the substrate.
• The active site binds the substrate forming
enzyme-substrate (ES) complex. This ES is
converted to enzyme-product (EP) which
dissociated to enzyme and product.

•
2. Catalytic efficiency:
• Most enzymes of high efficient in catalytic
activity reaching from 10
3
to 10
8
faster than
un catalyzed reactions. Each enzyme
molecule is capable of transforming 100 to
1000 substrate molecules into product each
second. So,
turnover number
is the number of
molecules of substrate converted to product
per enzyme molecule per second.

•
3. Specificity:
• Enzymes are high specific by interaction with one
or few specific substrates and catalyzing one type
of chemical reaction.
•
4. Cofactors:
• Some enzymes needed a non protein cofactors for
their enzymatic activity. These cofactors include
metal ions as Zn
2+
, Fe
2+
and organic molecules,
known as coenzymes, that are often vitamins
derivatives as NAD
+
, FAD and coenzyme A.

• Holoenzyme: enzyme with its cofactor.
• Apoenzyme: protein portion of the holoenzyme.
No biological activity of apoenzyme in case of
absence of appropriate cofactors.
• Prosthetic group: it is tightly bound coenzyme that
does not dissociate from the enzyme, ex. Biotin of
carboxylases.
• Enzyme activity can be regulated as activated or
inhibited, so, the rate of product formation
responds to the needs.

• Many enzymes are localized in
specific organelles within the cell.
This will lead to isolate the reaction
substrate or product from other
competing reactions, providing good
environment for reaction and to
regulate the thousands of enzymes in
the cells into their purpose pathways.

•
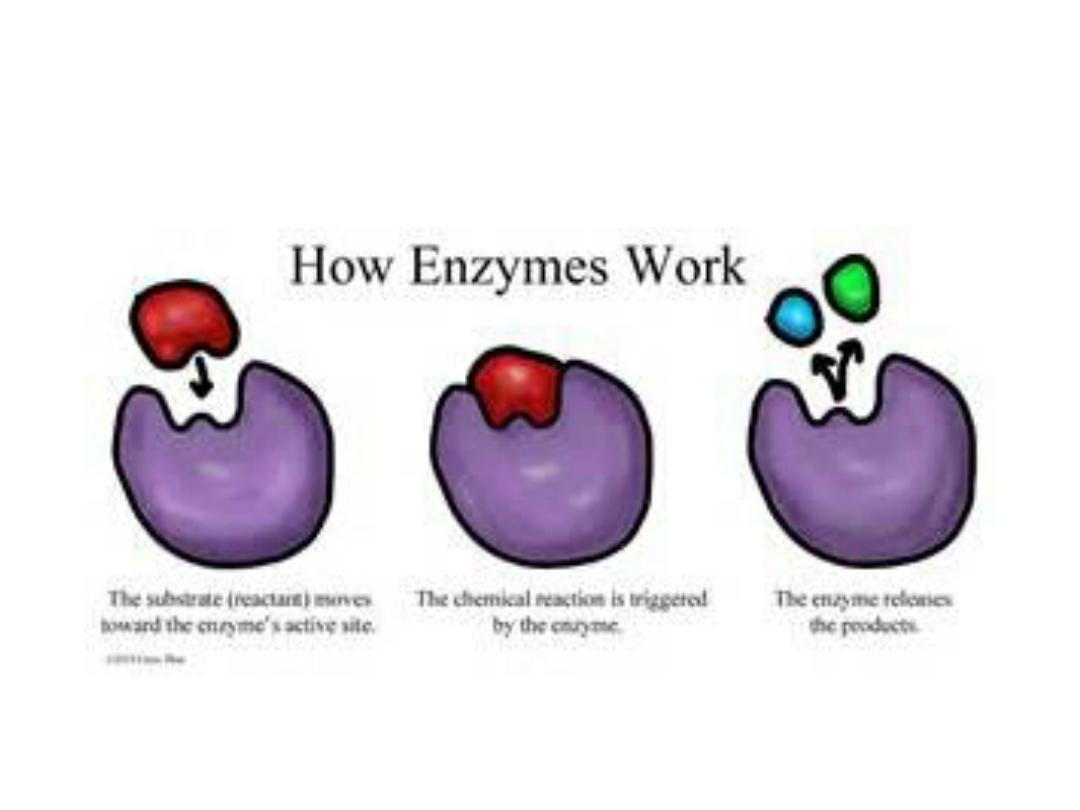
How enzyme work:
• All enzymes are proteins, but not all proteins
are enzymes. They are a biological catalysts,
mediate the synthesis of biological
compounds and catalyze reactions that
supply the cells with energy.
• Therefore, enzymes are responsible for all
the chemical reactions in the cells at which
covalent bonds are formed or broken.

• Enzymes act by lowering the activation
energy of the reaction they catalyze.
•
The action of enzymes have two faces:
• 1.Catalysis in terms of energy changes
which occur during the reaction. Enzymes
provide an alternate energetically
favorable reaction pathway different from
un catalyzed reaction.
• 2. The property of the active site
chemically to facilitate catalytic reaction
of the enzyme.

•
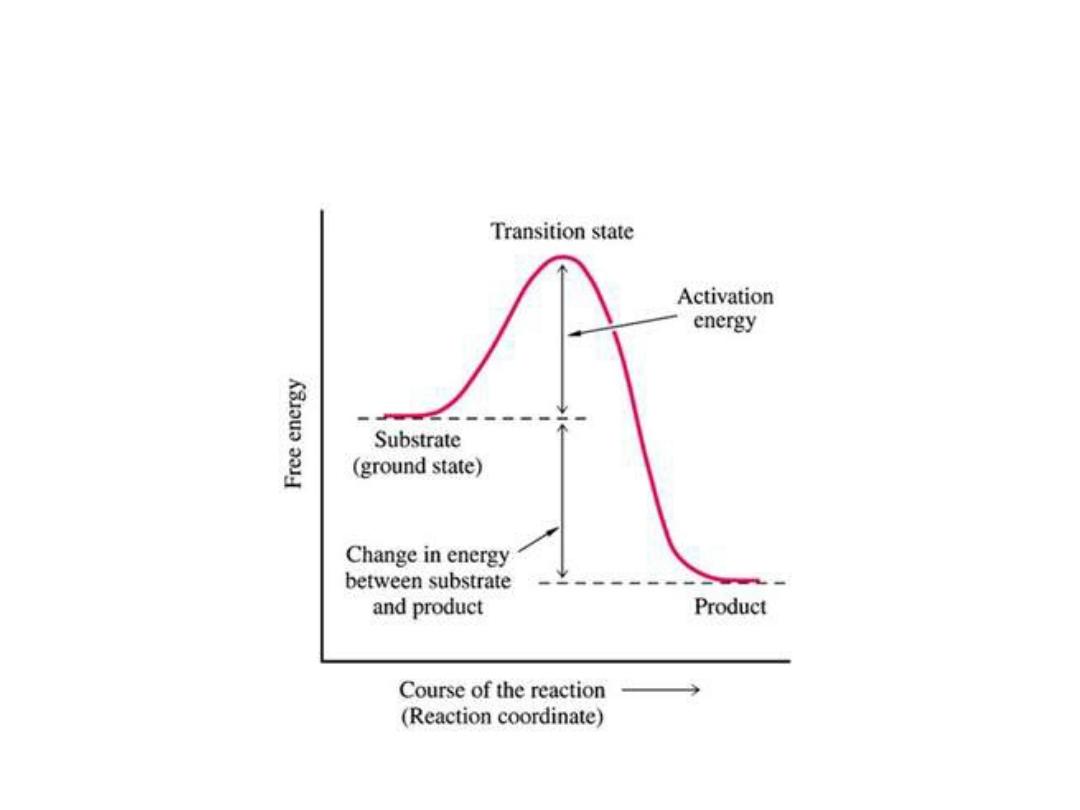
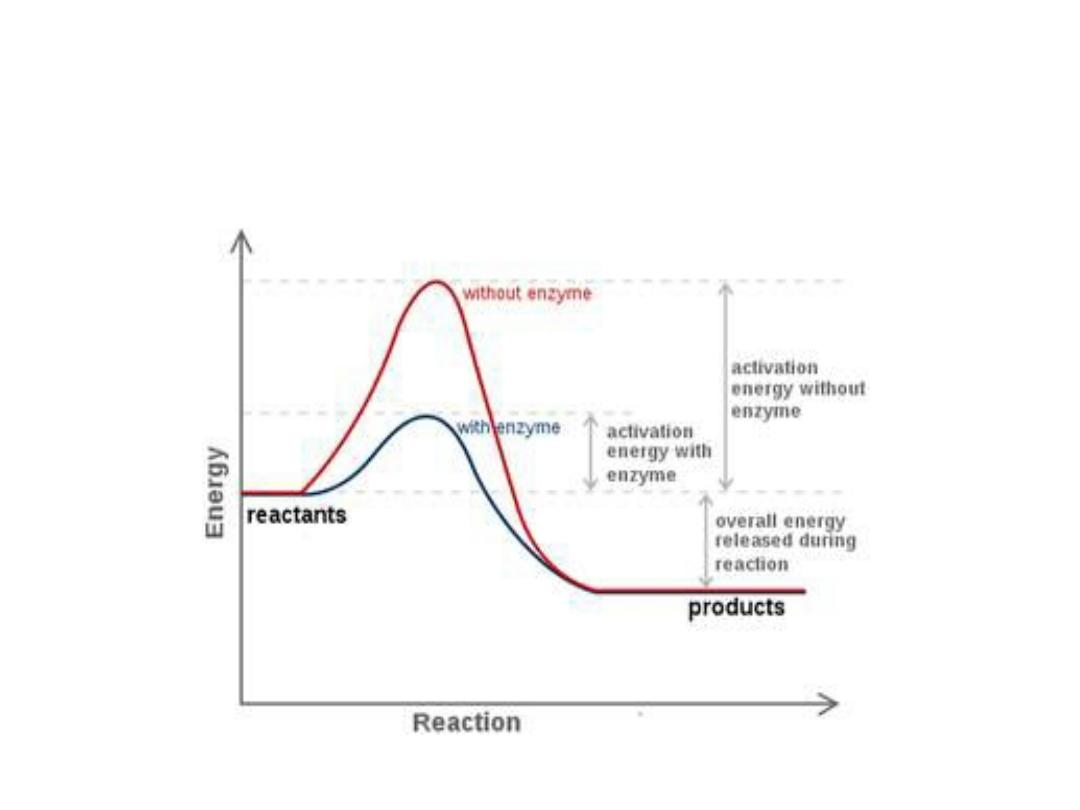
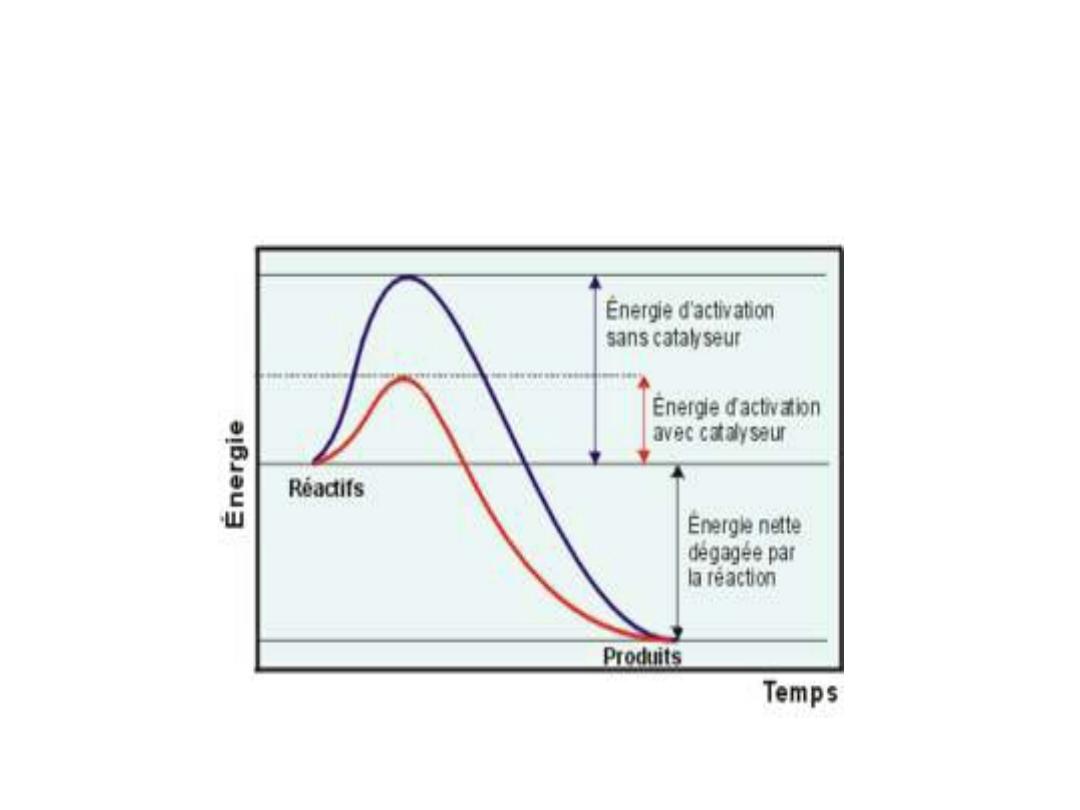
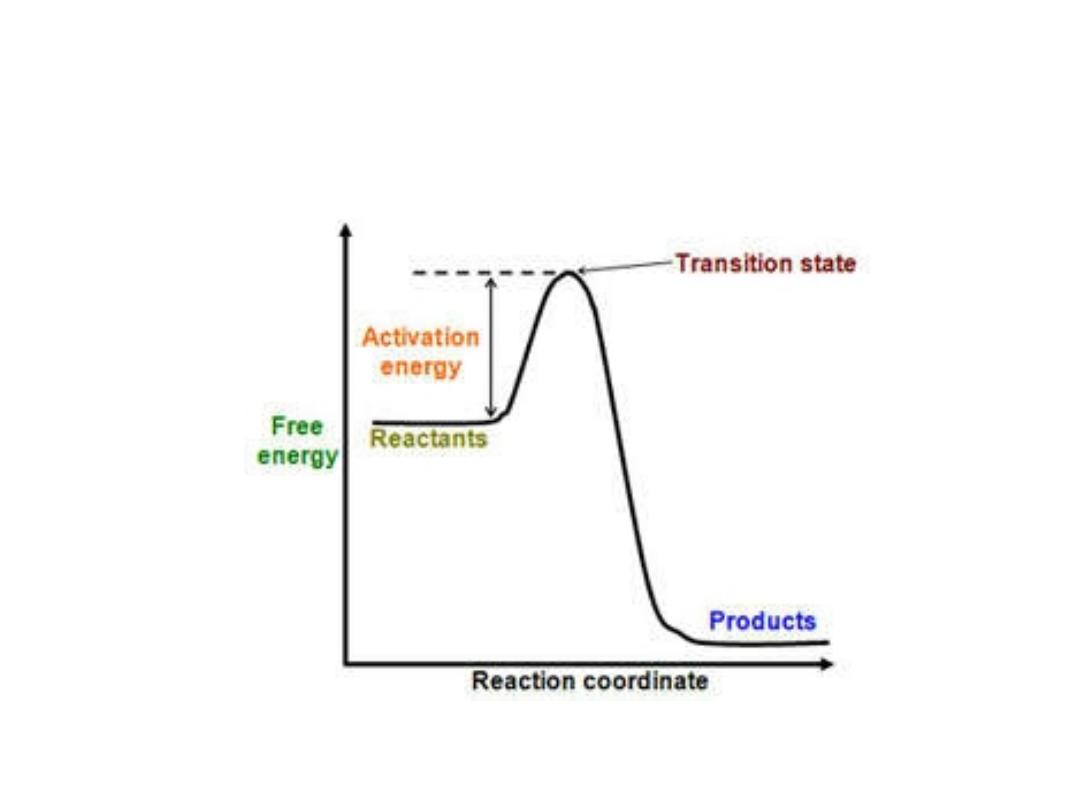
Energy changes during the reaction:
• For all the chemical reactions their will be energy
barrier separating the reactants and the products,
this is called
free energy of activation
. It is the
energy difference between the energy of the
reactants and the high energy intermediate that
occurs during the formation of products.
• A (substrate)
↔ T* ↔ B (product)
• E + S ---
→ ES ---→ EP ---→ P
• The changes in the energy during conversion of a
molecule of reactant A producing B through
transition state T*
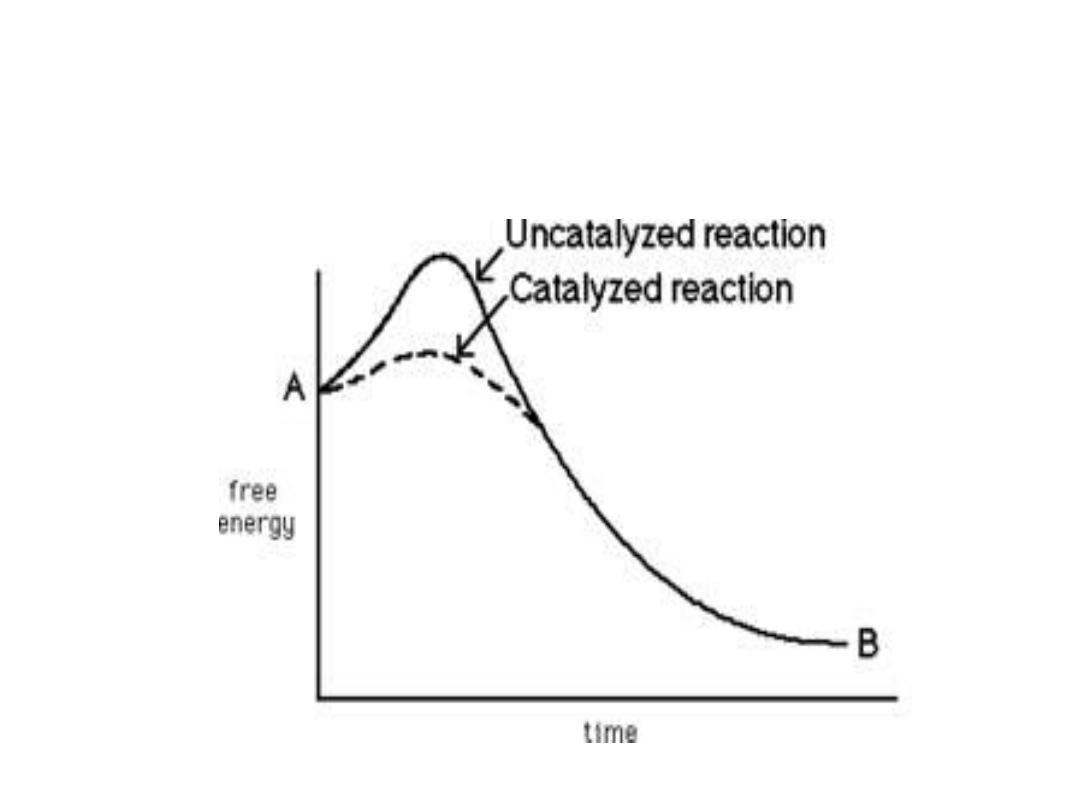
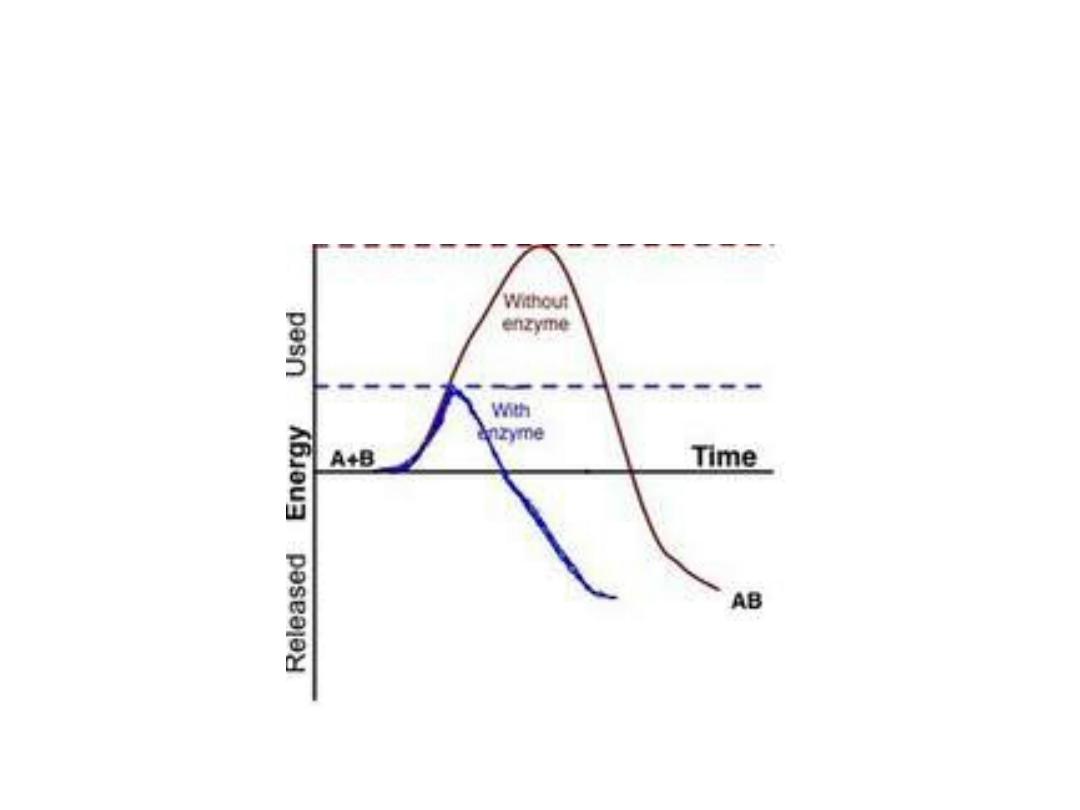

•
Effect of enzyme on the activation
energy of reaction:
•
1. Free energy of activation:
• The peak energy represent the transition
state in which a high energy intermediate
is formed during the conversion of
reactant A to product B. Due to the large
activation energy, the rate of un catalyzed
chemical reaction are often slow.

•
2. Rate of reaction:
• The molecules to be react, they must have
sufficient energy to overcome the energy
barrier of the transition state. So, in the
absence of an enzyme, only a small
amount of molecules between the
reactant and product may have enough
energy to reach the transition state

• The rate reaction is decided by the number of
energized molecules. Generally, the lower the free
energy of activation, the more molecules have
sufficient energy to pass over the transition state,
and thus, the faster the rate of the reaction.
•
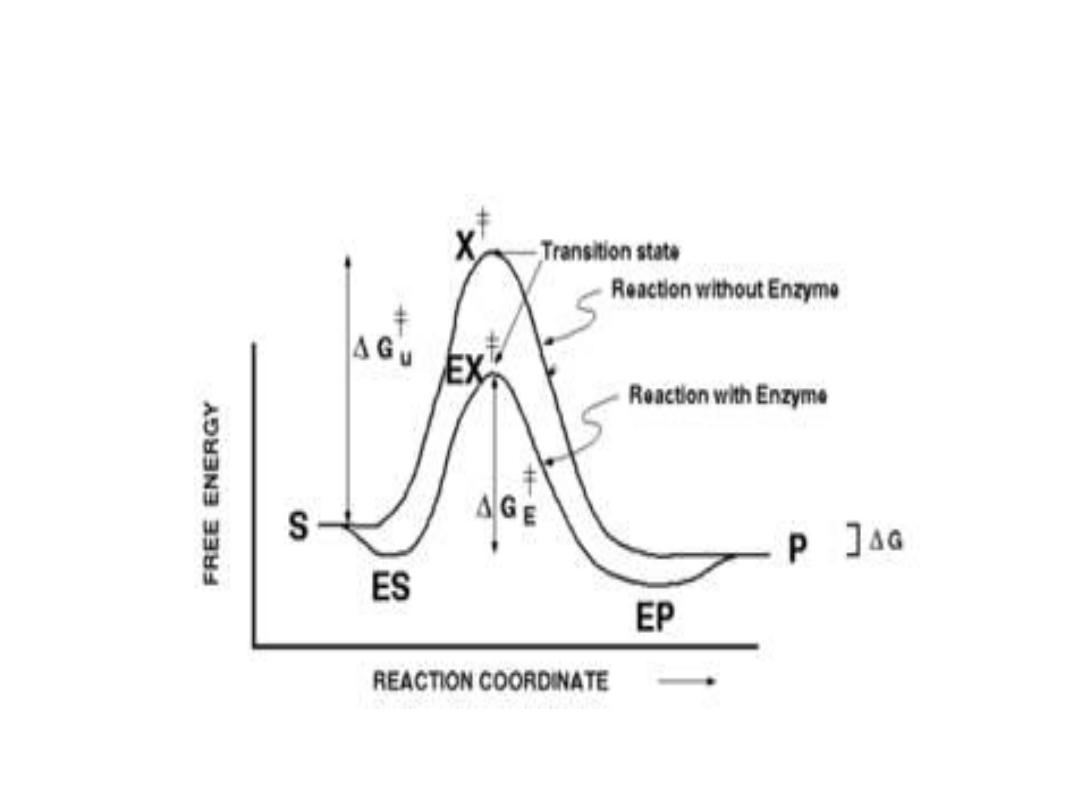
3. Alternate reaction pathway:
• Enzyme allows to proceed reaction rapidly in the
cell by alternate reaction pathway with a lower free
energy of activation. The enzyme does not change
the free energy of reactants or products and does
not change the equilibrium of reaction.

• Chemistry of the active site:
• Active site is a complex molecular machine posses
a diversity of chemical mechanisms to facilitate
the conversion of substrate to product.
•
Factors responsible for the catalytic efficiency of
enzymes:
•
1. Transition state stabilization:
• The active site acts as a flexible molecular
template that binds the substrate in a geometry
structurally resembling the activated transition
state of the molecule (T*).

• By stabilizing the substrate in its transition
state, the enzyme greatly increases the
concentration of the active intermediate
which converted to product, so accelerates
the reaction.
•
2. The active site provide catalytic groups
which enhance the formation of transition
state:
Some time active site involve in general
acid-base catalysis when the amino acid
residues provide or accept protons.

•
3. Visualization of transition state:
• Enzymes catalyzed conversion of
substrates to products. This spend high
energy of activation, first enzyme come in
contact with the substrate (ES), then
guiding the reaction analogous to the ES
transition state. The ES present at lower
energy than unbound substrate(S).

• Enzyme represent a parent who need to
remove sweater from his or her infant who is
unable to do that. The parent come in
contact with the infant as (ES), guiding the
infant’s arms into extended in vertical
position, which facilitates the removal of the
sweater, forming the disrobed baby, which
represent product. This will lead to spend
energy, so, substrate bound enzyme have
slightly low energy than unbound substrate
(S).

•
Factors affecting reaction velocity:
• Different enzymes shows different response
to changes in substrate concentration;
temperature and pH.
•
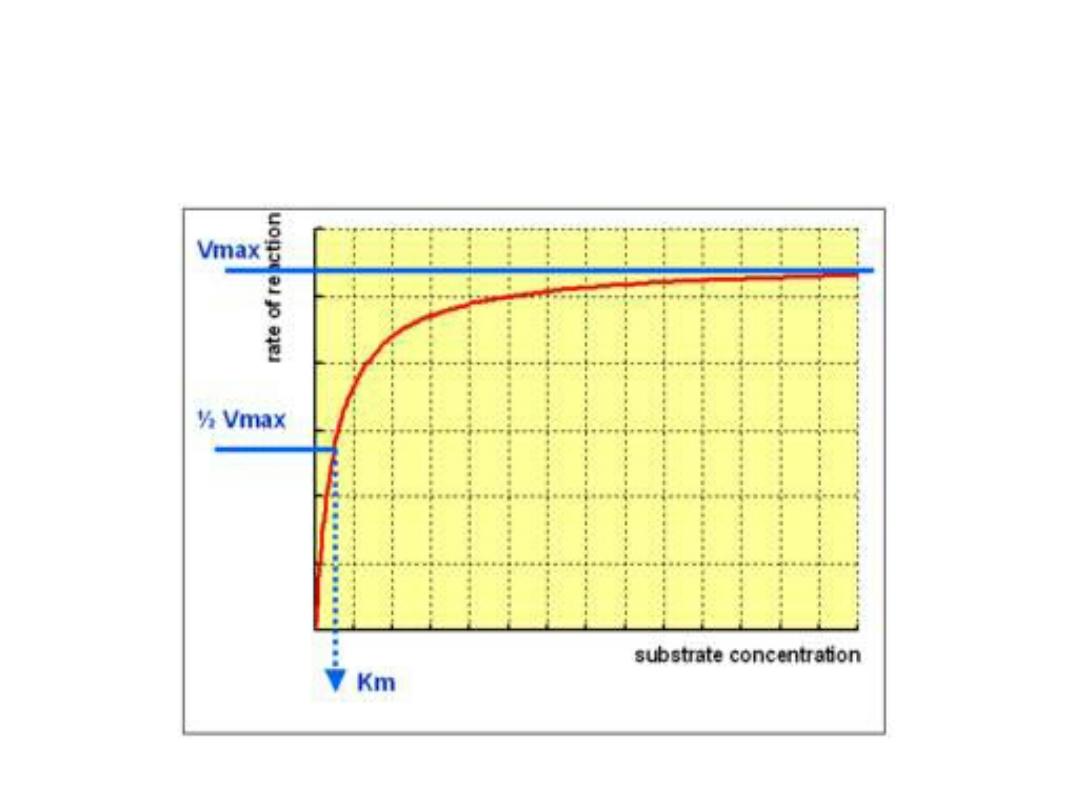
1. Substrate concentration:
•
The rate or velocity of a reaction (V)
is the
number of substrate molecules converted to
product per unit of time.
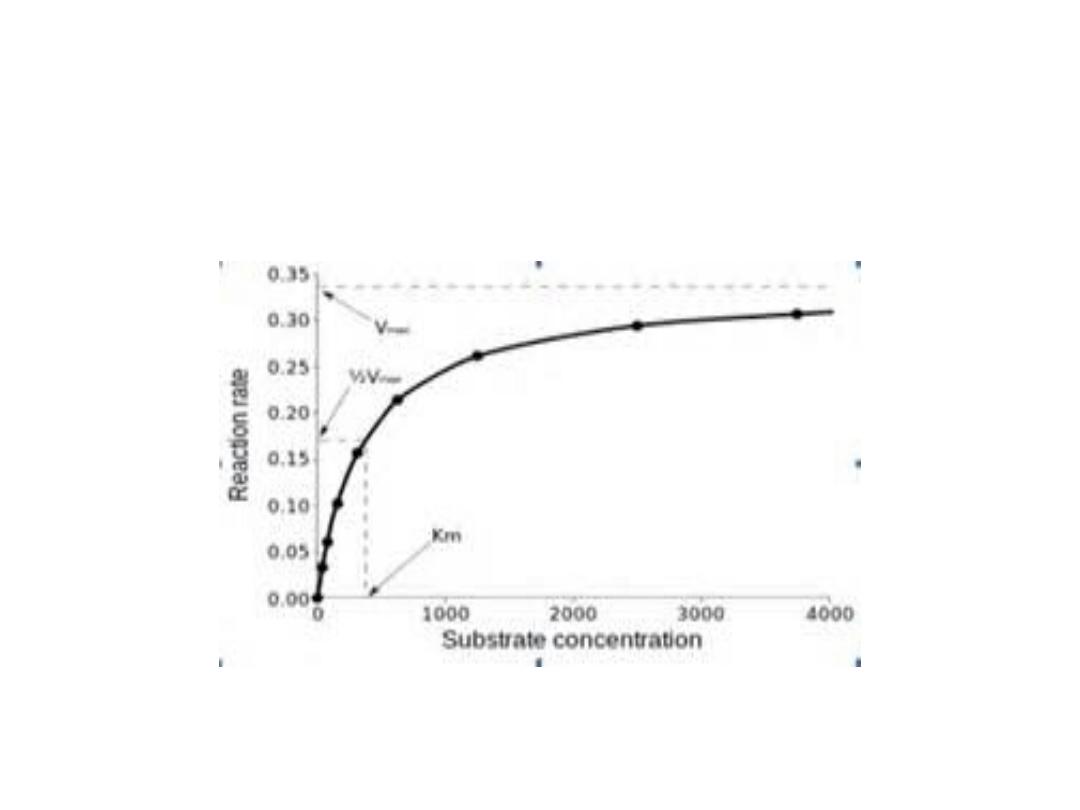

• It is expressed as µmoles product formed
per minute. The rate of enzyme reaction
increases with substrate concentration
until reach a maximal velocity (V
max
).
• The reaction velocity off at high substrate
concentration, and this reflects the
saturation with substrate of all available
binding sites on the enzyme.

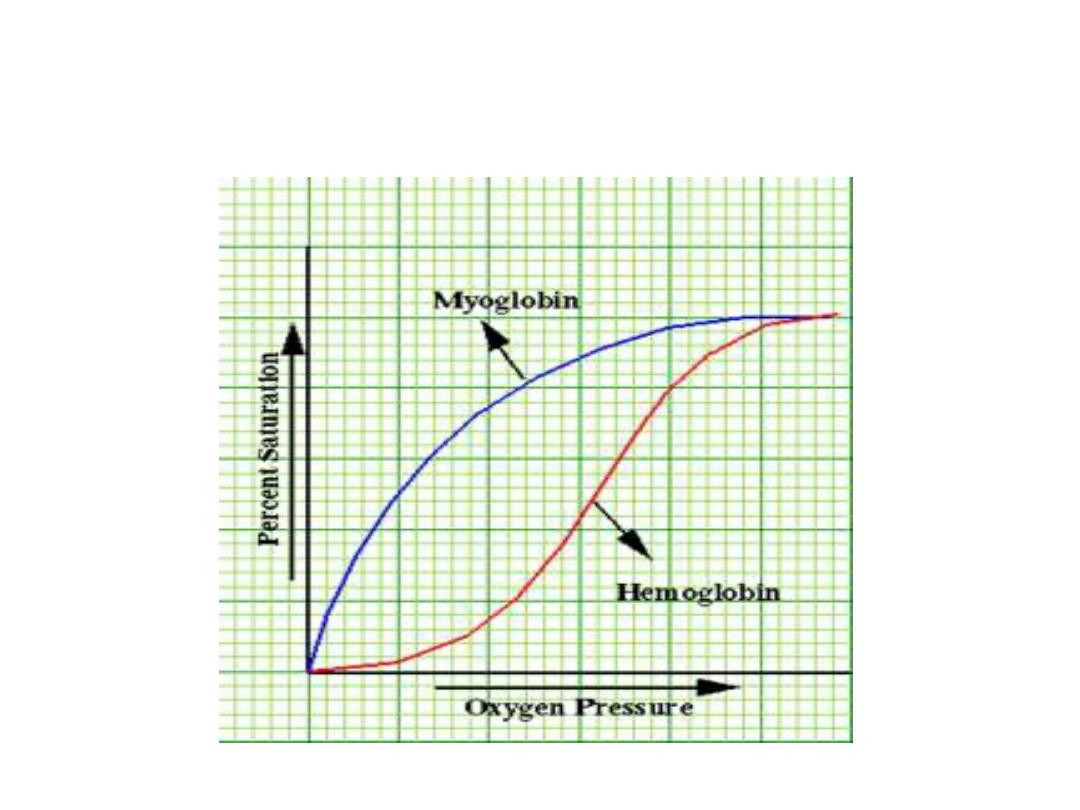
• Most enzymes shows hyperbolic shape
similar to oxygen dissociation curve of
myoglobin when draw reaction velocity
against substrate concentration. While
allosteric enzymes shows sigmoid curve
similar in shape to the oxygen
dissociation curve of hemoglobin.
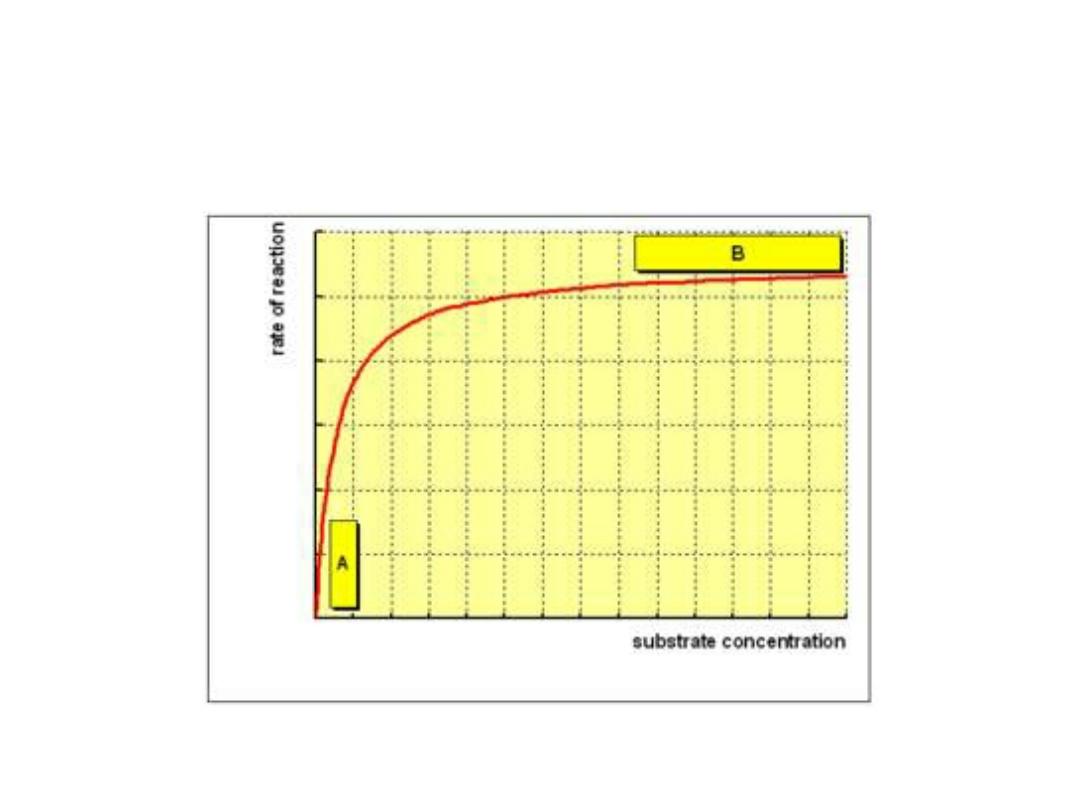

•
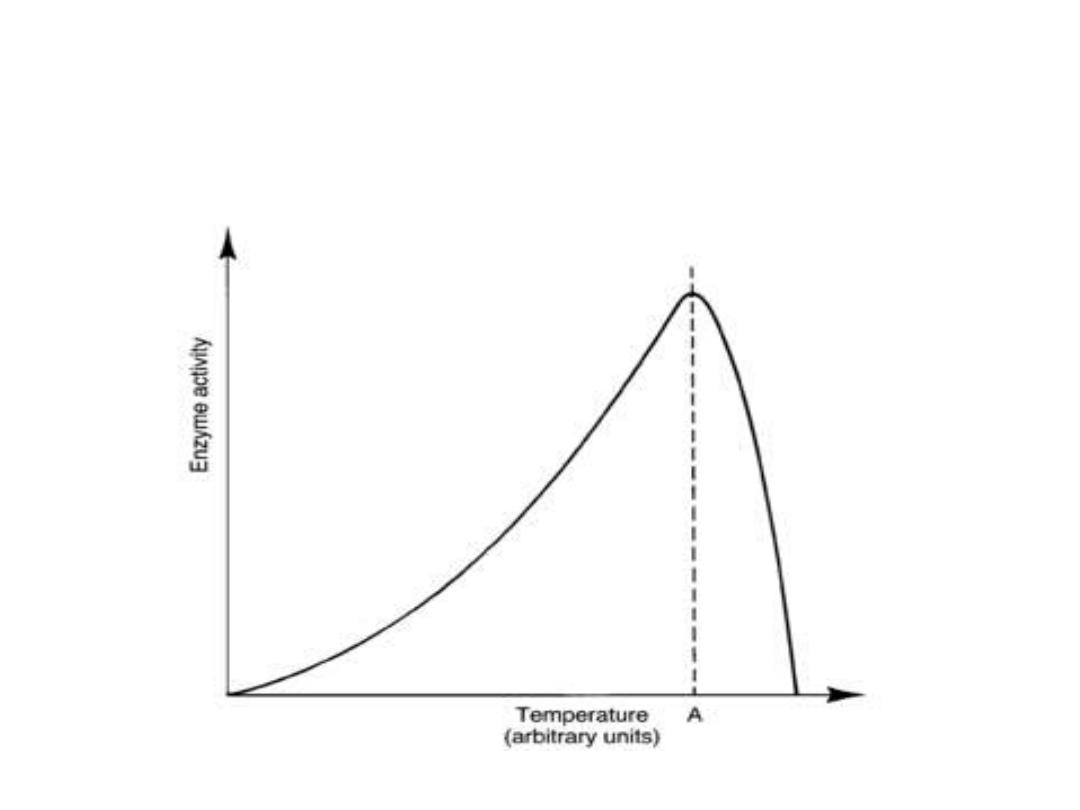
2. Temperature:
• Reaction velocity increases with temperature
until a peak of velocity is increased. This
increase occur due to increasing number of
molecules which have sufficient energy to pass
the energy barrier forming the product. More
increasing in temperature result in a decrease
in reaction velocity, as more temperature will
reduce denaturation of the enzyme.
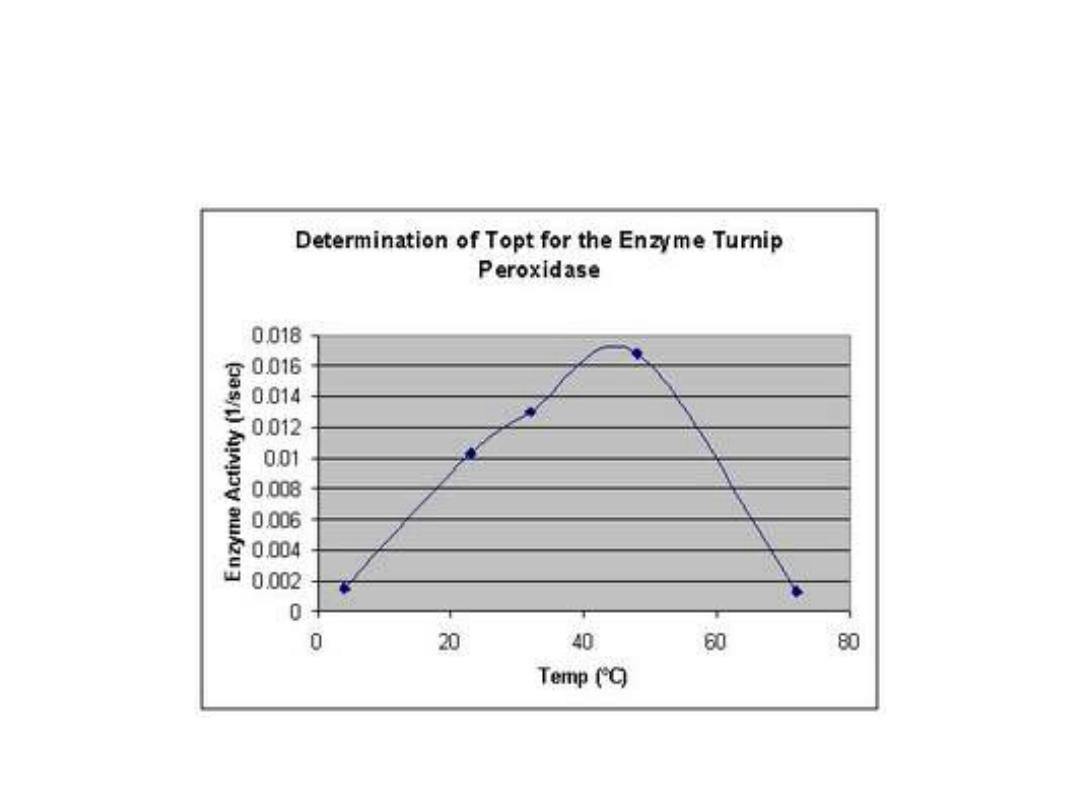

•
3. pH:
• Effect of pH on ionization of active site:
• Hydrogen ion concentration [H
+
] affects reaction
velocity in several ways, as the catalytic activity of
an enzyme requires the enzyme and substrate
should have specific ionized or unionized chemical
groups inorder to interact; ex: The catalytic activity
require the amino group of the enzyme in a
protonated form (-NH
3
+
). At alkaline pH this group
is deprotonated, so, the rate of reaction therefore
declines.

• Extremes of pH will lead to denaturation of
the enzyme due to the structure of the
catalytic active protein molecule depends on
the amino acid character of the side chains.
The maximal enzyme activity reached at
different pH and differ at different enzymes
and this is reflect the [H
+
] at which the
enzyme function in the body, ex: pepsin, the
digestive enzyme in the stomach maximally
active at pH 2. Some designed to work at
neutral pH which denaturized at high acidic
pH.
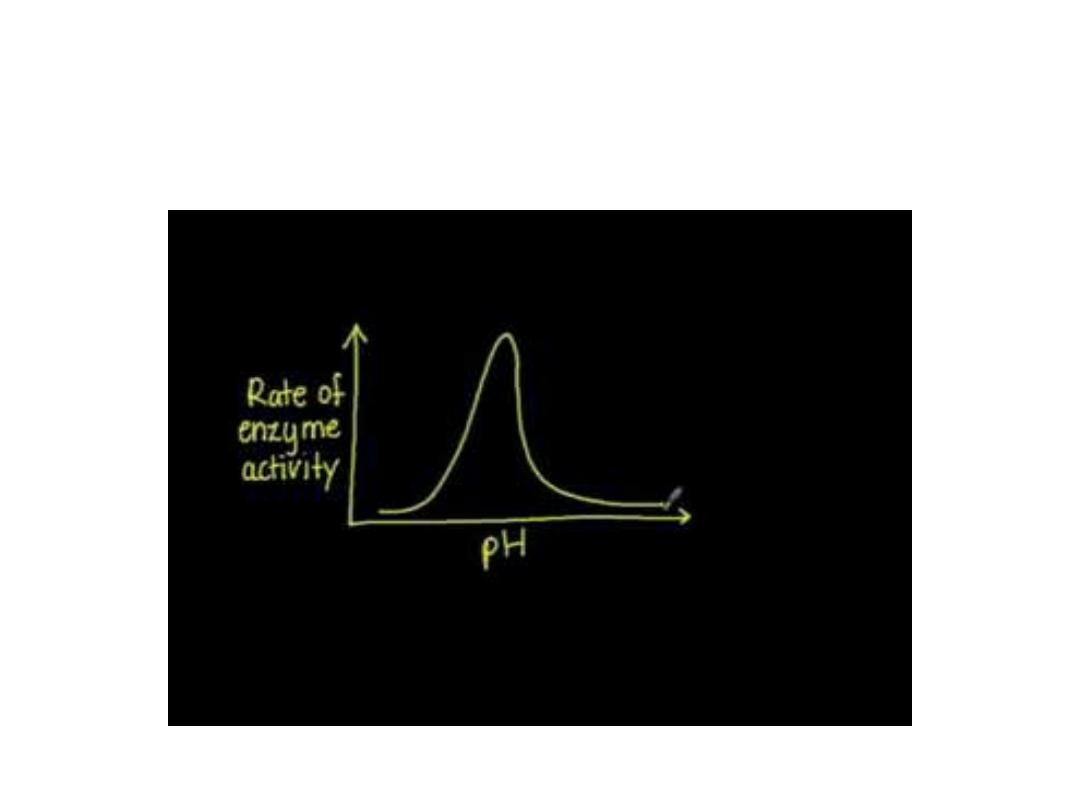
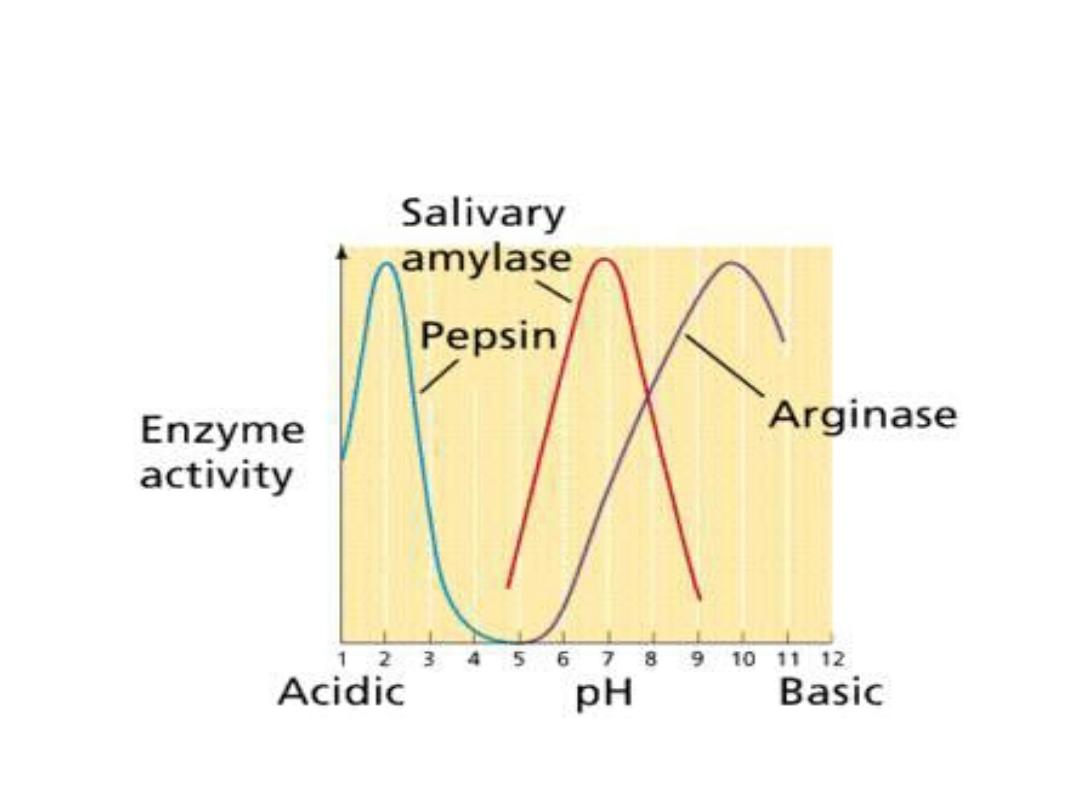
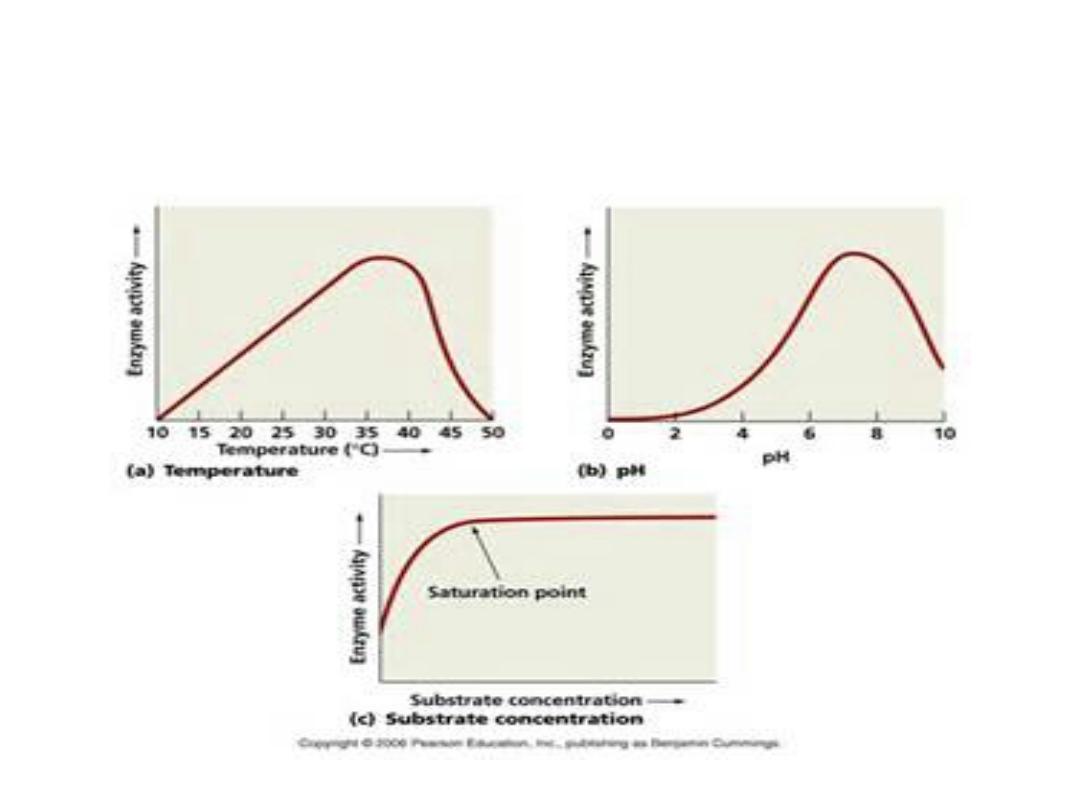

•
Enzyme unit:
• The amount of an enzyme that will catalyze the
transformation of a specified amount of substrate
into product under defined conditions.
• International unit (I.U) of enzyme corresponds to
the amount of enzyme that causes the loss of 1
µmole of substrate/ minute under specified
conditions, usually a saturating condition of
substrate.
• Katal (kat): The amount of enzyme that cause the
loss of one mole of substrate/ second Under
specified condition.

•
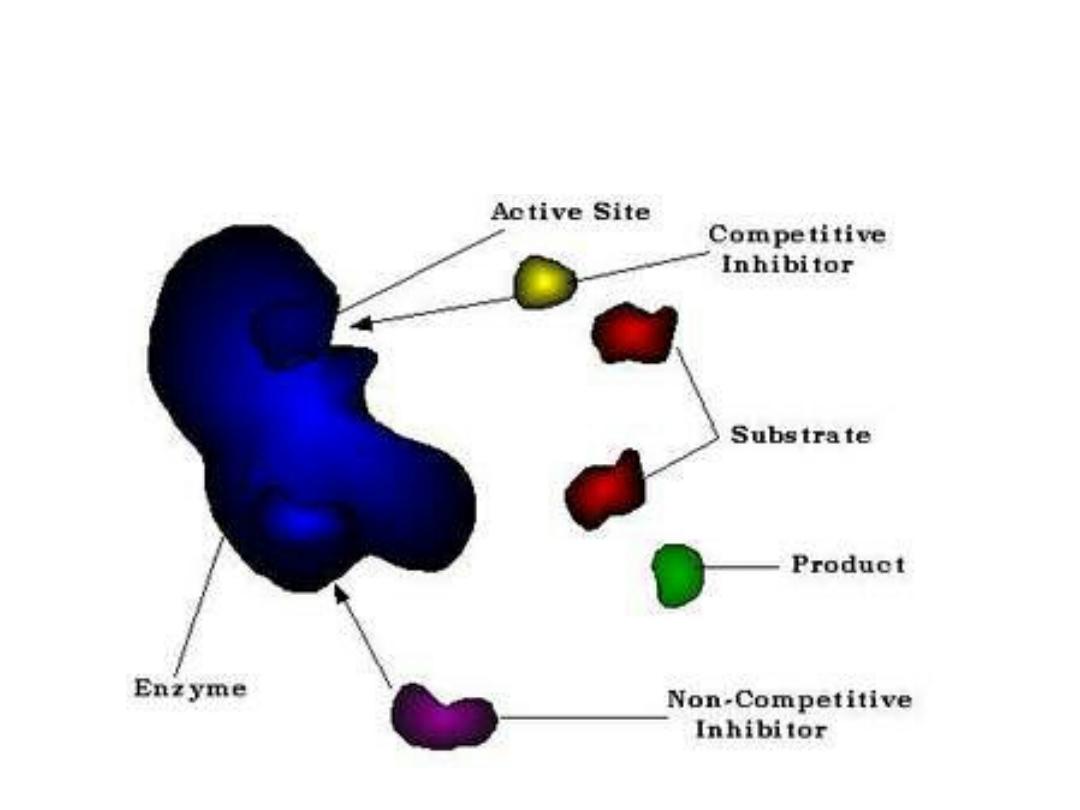
Enzyme Inhibitors:
• A great variety of naturally occurring and
synthetic compounds have the ability to
bind reversibly or irreversibly to specific
enzymes and alter their activity.
• Enzyme inhibitors reduce or eliminate the
catalytic activity of the enzymes.
• Ex: Drugs, Antibiotics, Toxins and
Antimetabolites.

•
Two general classes of inhibitors:
• 1.
Irreversible inhibitors:
• Form covalent bonds with specific functional
groups, amino acid side chain.
• Can not be released by dilution or dialysis, its
effects can not be reversed simply by increasing
the [S].
• Occurs when the inhibited enzyme does not regain
activity upon dilution of the enzyme- inhibitor
complex. Ex: The neurotoxic effects of certain
insecticides are due to their irreversible binding at
the catalytic site of the enzyme acetylcholine
esterase.

• 2.
Reversible inhibitors (RI):
• Binding to enzymes through non covalent
bonds.
• Dilution of the enzyme inhibitor complex
result in dissociation of the reversibly-
bound inhibitor and recovery of enzyme
activity.
• Have only a transient association with the
enzyme.

•
Two important types of inhibition:
•
1.Competitive inhibition (CI):
• In CI compounds that may or may not be
structurally related to the natural substrate
combine reversibly with the enzyme at or
near the active site.
• Both the substrate and the inhibitor
compete for the same site which leads to
a series of reactions.

• E + S
↔ ES → E + P
+
I
k
1
↕k
2
EI
(inactive)
K
I
= k
2
/k
1
(k
I
is dissociation constant for
the EI complex).

• In CI, ES and EI complexes are formed but EIS
complexes never produced.
• In conclusion, the high concentrations of substrates
will overcome the inhibition by shifting the reaction to
the right.
COOH COOH
ɪ ɪ
CH2 2H CH
ɪ
←--↗--→ ɪ
CH2 HC
ɪ ɪ
COOH COOH
Succinic acid Fumaric acid

Malonic acid is a (CI) of the enzyme
succinic dehydrogenas
COOH
ɪ
CH2
ɪ
COOH

• Ex. of (CI), succinate dehydrogenase enzyme
catalyzes the oxidation of succinate to
fumarate. Malonate is structurally similar to
the substrate (succinate) and competes for
binding at the active site of the enzyme.
Increasing succinate concentration, the
probability that the active site is occupied by
the substrate molecules rather than an
inhibitor.

•
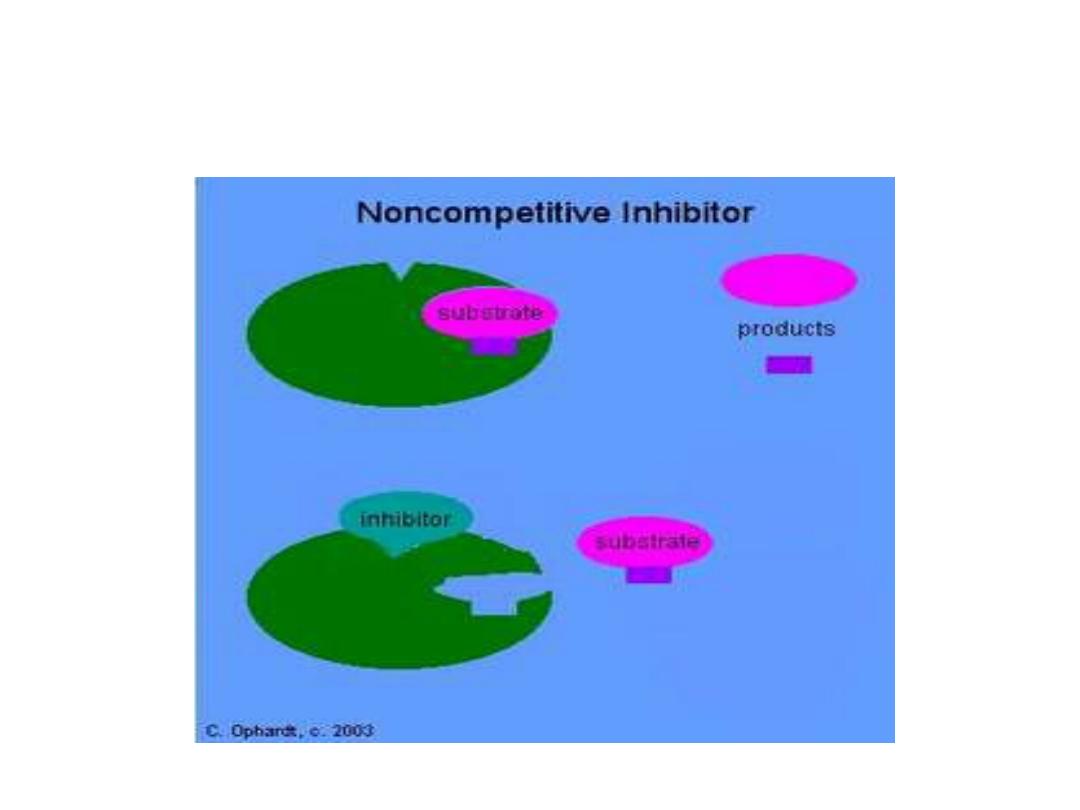
2. Non competitive inhibition (NCI):
• Compounds that are reversibly bind with
either the enzyme or the enzyme substrate
complex are designated as NCI.
• E + S
↔ ES → E + P
+ +
I I
↕ ↕
EI
↔ ESI
↘ (inactive)↙

• Ex: Reagent that can combine reversibly
with the –SH groups of cystein residues
that are essential for the catalytic activity
of some enzymes. ex. (Cu
2+
, Hg
2+
and
Ag
+
).
• E-SH + Ag
+
↔ E-S-Ag + H
+
• This type of inhibition is not completely
reversed by high [S], since the cyclic
sequence of the above equation will occur
regardless of the [S].

• NCI occurs when the inhibitor and
substrate bind at different site on the
enzyme.
• NCI can bind either free enzyme or ES
complex so prevent occurring of reaction.
• NCI can not be overcome by increasing
the substrate concentration.
• NCI does not interfere with the binding of
substrate to enzyme.

• Many drugs act as inhibitors of enzyme, Ex. Many
antibiotics as penicillin and amoxicillin, act as
inhibiting one or more of the enzymes of bacterial
cell wall synthesis.
• Drugs may also act by inhibiting extracellular
reactions as angiotensin converting enzyme (ACE)
inhibitors, which lower blood pressure by blocking
the enzyme that cleaves angiotensin 1 to form the
potent vasoconstrictor angiotensin 11. These
drugs as captopril, enalopril and lisinopril cause
vasodilation and reduce blood pressure.

•
Uncompetitive inhibition (UCI):
• Compounds that reversibly combine with only
ES complex, but not the free enzyme are
called (UCI). The inhibition is not overcome
by high substrate concentration, Ex:
• Succinate + CoA + GT
↔ Succinyl CoA +
GDP + Pi
• Enzyme: Succinyl CoA synthetase.
• Pi is the UCI

•
Enzyme theories proposed to explain
the specificity of enzyme action:
•
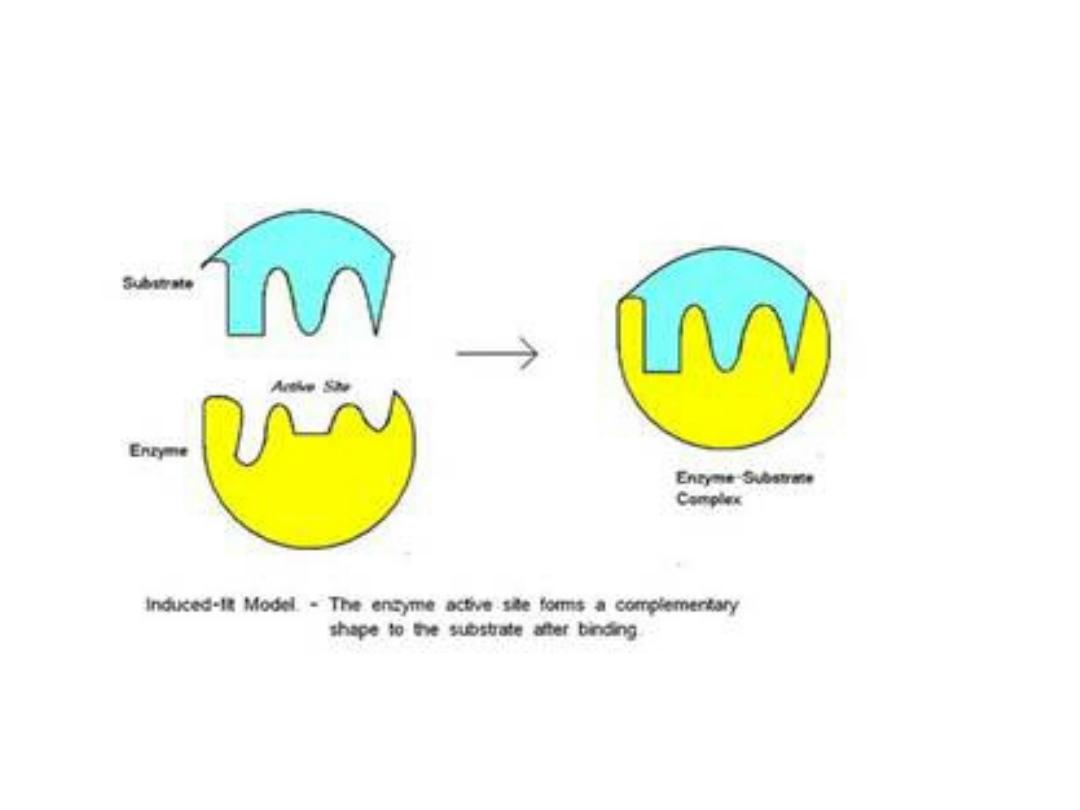
1. Lock and key theory:
• The active site of the enzyme is
complementary in conformation to the
substrate, so that enzyme and substrate
recognize one another.

•
2. Induced-fit theory:
• The enzyme changes its shape upon
binding the substrate, so, the
conformation of substrate are only
complementary after the binding reaction.