DNA Replication
Genetic information is transmitted from parent to progeny by replication of parental DNA, a process in which two daughter DNA molecules are produced that are each identical to the parental DNA molecule.Replication:
The overall process of DNA replication in prokaryotes and eukaryotes is compared
The bacterial chromosome is a closed, double-stranded circular DNA molecule having a single origin of replication. Separation of the two parental strands of DNA creates two replication forks that move away from each other in opposite directions around the circle. Replication is, thus, a bidirectional process. The two
replication forks eventually meet, resulting in the production of two identical circular molecules of DNA.
Each eukaryotic chromosome contains one linear molecule of dsDNA having multiple origins of replication. Bidirectional replication occurs by means of a pair
of replication forks produced at each origin. Completion of the process results in
the production of two identical linear molecules of dsDNA (sister chromatids). DNA replication occurs in the nucleus during the S phase of the eukaryotic cell
cycle. The two identical sister chromatids are separated from each other when the cell divides during mitosis. The structure of a representative eukaryotic chromosome during the cell cycle is Shown below
Eukaryotic Chromosome Replication During S-Phase
COMPARISON OF DNA AND RNA SYNTHESISThe overall process of DNA replication requires the synthesis of both DNA and RNA. These two types of nucleic acids are synthesized by DNA polymerases and
RNA polymerases, respectively. DNA synthesis and RNA synthesis are compared
Similarities include:
• The newly synthesized strand is made in the 5' →3' direction.
• The template strand is scanned in the 3' →5' direction.
• The newly synthesized strand is complementary and antiparallel to the
template strand.
• Each new nucleotide is added when the 3' hydroxyl group of the growing
strand reacts with a nucleoside triphosphate, which is base-paired with the template strand. Pyrophosphate (PPi, the last two phosphates) is released during this reaction. Differences include:
• The substrates for DNA synthesis are the dNTPs, whereas the substrates
for RNA synthesis are the NTPs.
• DNA contains thymine, whereas RNA contains uracil.
• DNA polymerases require a primer, whereas RNA polymerases do not.
That is, DNA polymerases cannot initiate strand synthesis, whereas RNA
polymerases can.
• DNA polymerases can correct mistakes ("proofreading"), whereas RNA
polymerases cannot. DNA polymerases have 3' → 5' exonuclease activity
for proofreading.
STEPS OF DNA REPLICATION
The molecular mechanism of DNA replication is shown below .The sequence of events is as follows:
1. The base sequence at the origin of replication is recognized.
2. Helicase breaks the hydrogen bonds holding the base pairs together. This
allows the two parental strands of DNA to begin unwinding and forms two replication forks.
3. Single-stranded DNA binding protein ( SSB) binds to the single-stranded
portion of each DNA strand, preventing them from reassociating and protecting them from degradation by nucleases.
4. Primase synthesizes a short (about 10 nucleotides) RNA primer in the
5'→ 3'direction, beginning at the origin on each parental strand. The parental strand is used as a template for this process. RNA primers are required because DNA polymerases are unable to initiate synthesis of
DNA, and can only extend a strand from the 3' end of a preformed "primer."
5. DNA polymerase III begins synthesizing DNA in the 5'→3' direction, beginning at the 3' end of each RNA primer. The newly synthesized strand is complementary and antiparallel to the parental strand used as a template. This strand can be made continuously in one long piece and is known as the "leading strand:'
• The "lagging strand" is synthesized discontinuously as a series of small
fragments (about 1 ,000 nucleotides long) known as Okazaki fragments.
Each Okazaki fragment is initiated by the synthesis of an RNA primer by
primase, and then completed by the synthesis of DNA using DNA polymerase
III. Each fragment is made in the 5' to 3' direction.
• There is a leading and a lagging strand for each of the two replication
forks on the chromosome.
6. RNA primers are removed by RNAase H in eukaryotes and an uncharacterized
DNA polymerase fills in the gap with DNA. In prokaryotes DNA polymerase I both removes the primer (5' exonuclease) and synthesizes new DNA, beginning at the 3' end of the neighboring Okazaki fragment.
7. Both eukaryotic and prokaryotic DNA polymerases have the ability to "proofread" their work by means of a 3'→5' exonuclease activity. If DNA polymerase makes amistake during DNA synthesis, the resulting unpaired base at the 3' end of the growing strand is removed before synthesis continues.
8. DNA ligase seals the "nicks" between Okazaki fragments, converting them
to a continuous strand of DNA.
9. DNA gyrase (DNA topoisomerase II) provides a "swivel" in front of each replication fork. As helicase unwinds the DNA at the replication forks, the DNA ahead of it becomes overwound and positive supercoils form. DNA gyrase inserts negative supercoils by nicking both strands of DNA, passing the DNA strands through the nick, and then resealing both strands. Quinolones are a family of drugs that block the action of topoisomerases. Nalidixic acid kills bacteria by inhibiting DNA gyrase. Inhibitors of eukaryotic topoisomerase II (etoposide, teniposide) are becoming useful as anticancer agents.
The mechanism of replication in eukaryotes is believed to be very similar to this.
Eukaryotic DNA Polymerases
• DNA α and δ work together to synthesize both the leading and lagging strands.
• DNA polymerase Ɣ replicates mitochondrial DNA.• DNA polymerases β and ε are thought to participate primarily in DNA repair.
DNA polymerase ε may substitute for DNA polymerase δ in certain cases.
Telomerase
Telomeres are repetitive sequences at the ends of linear DNA molecules in eukaryotic chromosomes. With each round of replication in most normal cells, the telomeres are shortened because DNA polymerase cannot complete synthesis of the 5' end of each strand. This contributes to the aging of cells, because eventually the telomeres become so short that the chromosomes cannot function properly and the cells die.
Telomerase is an enzyme in eukaryotes used to maintain the telomeres. It contains a short RNA template complementary to the DNA telomere sequence, as well as telomerase reverse transcriptase activity (hTRT). Telomerase is thus able to replace telomere sequences that would otherwise be lost during replication. Normally telomerase activity is present only in embryonic cells, germ (reproductive) cells, and stem cells, but not in somatic cells. Cancer cells often have relatively high levels of telomerase, preventing the telomeres from becoming shortened and contributing to the immortality of malignant cells.
Telomerase
• Completes the replication of the telomere sequences at both ends of a eukaryotic chromosome
• Present in embryonic cells, fetal cells, and certain adult stem cells;not present in adult somatic cells
• In appropriately present in many cancer cells, contributing to their unlimited replication
Reverse Transcriptase
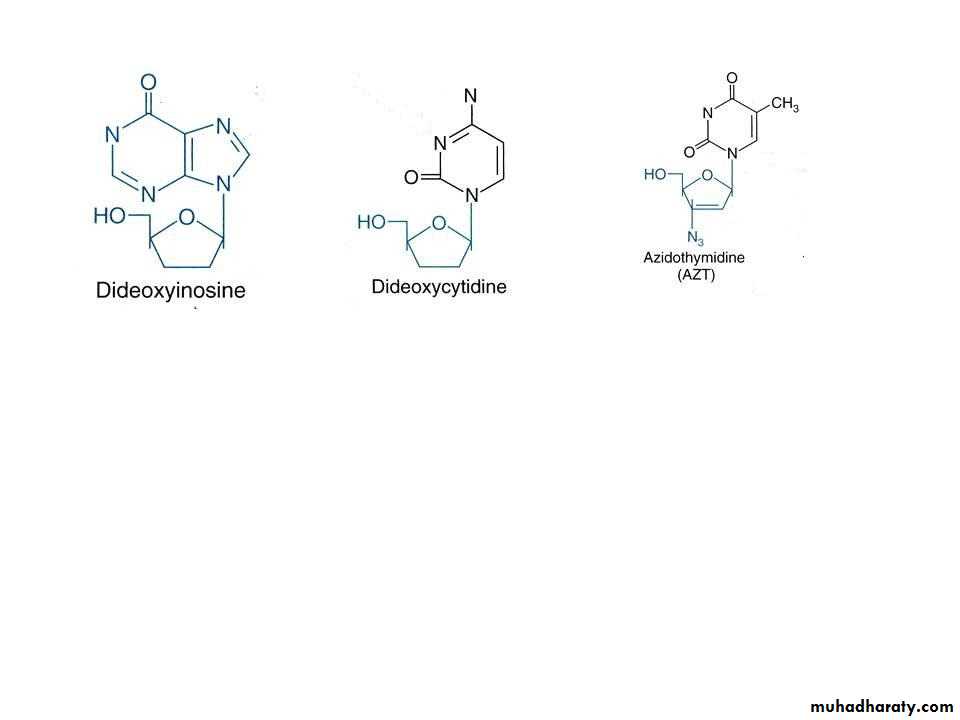
Reverse transcriptase is an RNA-dependent DNA polymerase that requires an RNA template to direct the synthesis of new DNA. Retroviruses, most notably HIV, use this enzyme to replicate their RNA genomes. DNA synthesis by reversetranscriptase in retroviruses can be inhibited by AZT, ddC, and ddl.
Quinolones and DNA Gyrase
Quinolones and fluoroquinolones inhibit DNA gyrase (prokaryotic topoisomerase II), preventing DNA replication and transcription. These drugs, which are most active against aerobic gram-negative bacteria, include:• Levofloxacin
• Ciprofloxacin
• Moxifloxacin
Resistance to the drugs has developed over time; current uses inclued treatment of gonorrhea and upper and lower urinary tract infections in both sexes.
Bridge to Pharmacology
One chemotherapeutic treatment of HIV is the use of AZT (3'-azido- 2', 3' -dideoxythymidine) or structurally related compounds. Once AZT enters cells, it can be converted to the triphosphate derivative and used as a substrate for the viral reverse transcriptase in synthesizing DNA from its RNA genome. The replacement of an azide instead of a norm al hyd roxyl group at the 3' position of the deoxyribose prevents further replication by effectively causing chain termination. Although it is a DNA polymerase, reverse transcriptase lacks proofreading activity.