Transcription
The synthesis of RNA molecules using DNA strands as the templates so that the genetic information can be transferred from DNA to RNA.Transcription
Both processes use DNA as the template.
Phosphodiester bonds are formed in both cases.Both synthesis directions are from 5´ to 3´.
Similarity between replication and transcription
• replication
• transcription• template
• double strands
• single strand
• substrate
• dNTP
• NTP• primer
• yes
• no• Enzyme
• DNA polymerase
• RNA polymerase• product
• dsDNA
• ssRNA• base pair
• A-T, G-C
• A-U, T-A, G-CDifferences between replication and transcription
• Template and Enzymes
The whole genome of DNA needs to be replicated, but only small portion of genome is transcribed in response to the development requirement, physiological need and environmental changes.
DNA regions that can be transcribed into RNA are called structural genes.
§1.1 Template
The template strand is the strand from which the RNA is actually transcribed.The coding strand is the strand whose base sequence specifies the amino acid sequence of the encoded protein.
• Only the template strand is used for the transcription, but the coding strand is not.
• Both strands can be used as the templates.• The transcription direction on different strands is opposite.
• This feature is referred to as the asymmetric transcription.
Asymmetric transcription
template strand
coding strandtemplate strand
coding strand§2 RNA Polymerase
• The enzyme responsible for the RNA synthesis is DNA-dependent RNA polymerase.
• The prokaryotic RNA polymerase is a multiple-subunit protein of ~480kD.
• Eukaryotic systems have three kinds of RNA polymerases, each of which is a multiple-subunit protein and responsible for transcription of different RNAs.
Holoenzyme
The holoenzyme of RNA-pol in E.coli consists of 5 different subunits: 2 .
2
• subunit
• MW• function
•
• 36512
• Determine the DNA to be transcribed
•
• 150618
• Catalyze polymerization
•
• 155613
• Bind & open DNA template
•
• 70263
• Recognize the promoter
• for synthesis initiation
RNA-pol of E. Coli
• Each transcriptable region is called operon.
• One operon includes several structural genes and upstream regulatory sequences (or regulatory regions).
• The promoter is the DNA sequence that RNA-pol can bind. It is the key point for the transcription control.
§3 Recognition of Origins
Promoter
Prokaryotic promoterConsensus sequence
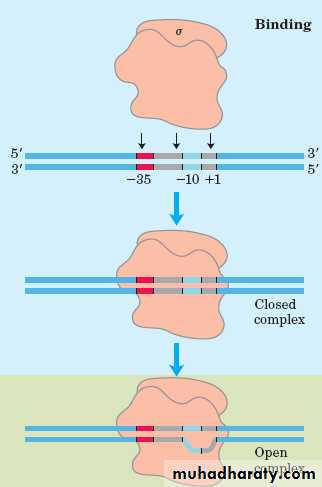
• The -35 region of TTGACA sequence is the recognition site and the binding site of RNA-pol.
• The -10 region of TATAAT is the region at which a stable complex of DNA and RNA-pol is formed.
• Section 2
• Transcription ProcessGeneral concepts
• Three phases: initiation, elongation, and termination.• The prokaryotic RNA-pol can bind to the DNA template directly in the transcription process.
• The eukaryotic RNA-pol requires co-factors to bind to the DNA template together in the transcription process.
§2.1 Transcription of Prokaryotes
Initiation phase: RNA-pol recognizes the promoter and starts the transcription.
Elongation phase: the RNA strand is continuously growing.
Termination phase: the RNA-pol stops synthesis and the nascent RNA is separated from the DNA template.
a. Initiation
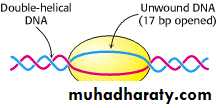
• RNA-pol recognizes the TTGACA region, and slides to the TATAAT region, then opens the DNA duplex.• The unwound region is about 171 bp.
The first nucleotide on RNA transcript is always purine triphosphate. GTP is more often than ATP.
The pppGpN-OH structure remains on the RNA transcript until the RNA synthesis is completed.
The three molecules form a transcription initiation complex.
RNA-pol (2) - DNA - pppGpN- OH 3
No primer is needed for RNA synthesis.
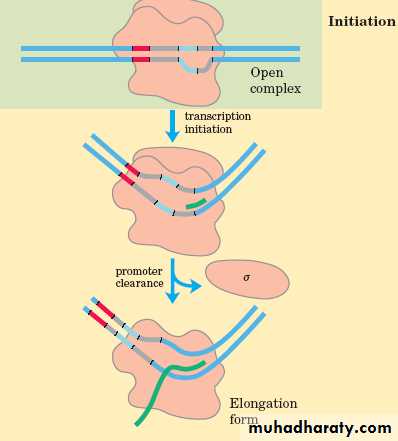
The subunit falls off from the RNA-pol once the first 3,5 phosphodiester bond is formed.The core enzyme moves along the DNA template to enter the elongation phase.
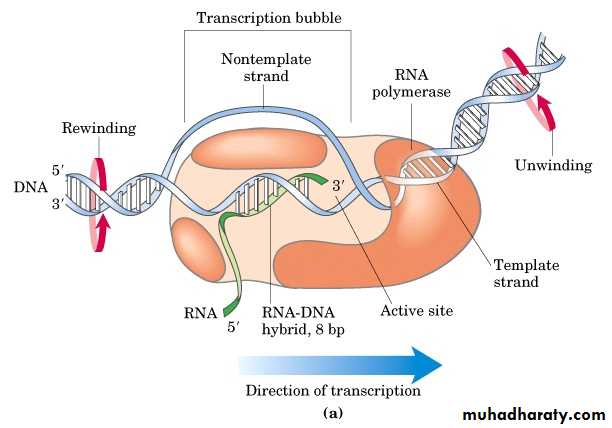
b. Elongation
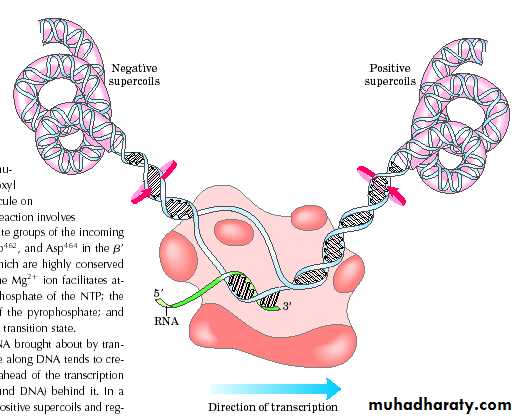
• The release of the subunit causes the conformational change of the core enzyme. The core enzyme slides on the DNA template toward the end.• Free NTPs are added sequentially to the 3 -OH of the nascent RNA strand.
.
• The 3 segment of the nascent RNA hybridizes with the DNA template, and its 5 end extends out the transcription bubble as the synthesis is processing.
Transcription bubble
RNA-pol of E. Coli
RNA-pol of E. Coli
•
•
c. Termination
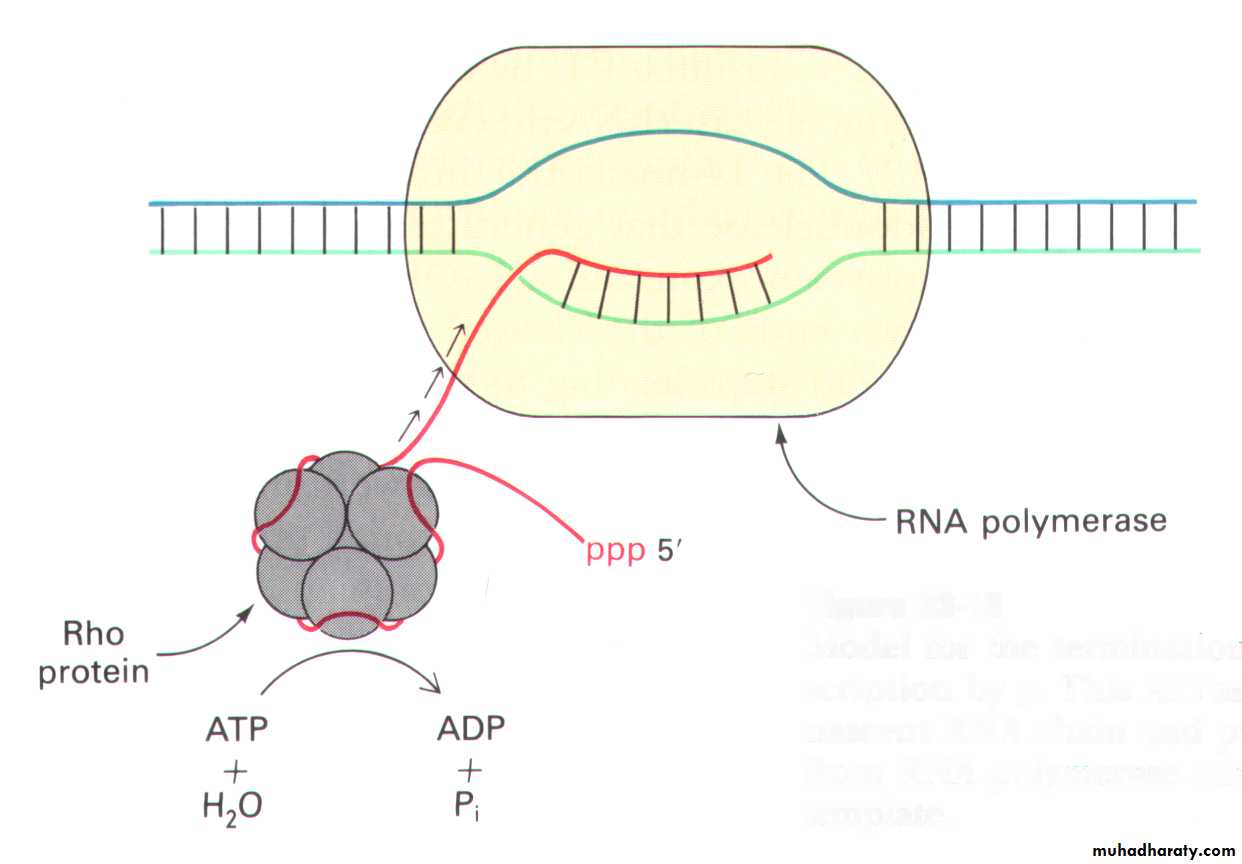
• The RNA-pol stops moving on the DNA template. The RNA transcript falls off from the transcription complex.• The termination occurs in either -dependent or -independent manner.
The termination function of factor
The factor, a hexamer, is a ATPase and a helicase.-independent termination
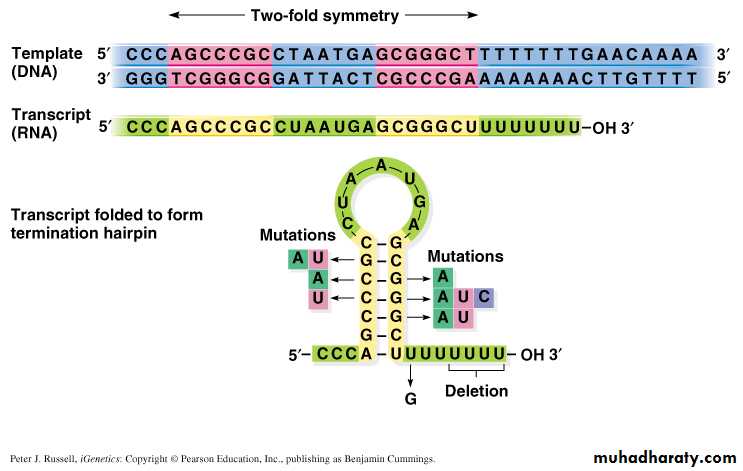
• The termination signal ( Palindrome) is a stretch of 30-40 nucleotides on the RNA transcript, consisting of many GC followed by a series of A.• The sequence specificity of this nascent RNA transcript will form particular stem-loop structures to terminate the transcription.
Fig. 5.5 Sequence of a -independent terminator and structure of the terminated RNA
The stem-loop structure alters the conformation of RNA-pol, leading to the pause of the RNA-pol moving.
Then the competition of the RNA-RNA hybrid and the DNA-DNA hybrid reduces the DNA-RNA hybrid stability, and causes the transcription complex dissociated.