DNA replication Laith Alghazaly
DNA function is information storage as a Sequence of strand stores on the gene. DNA passed to the descendant cells:Accurate copying
Repair of any damage to avoid changes
Accurate subdivision
DNA replication is the process that take place when cells divide , each new cell gets an exact copy of DNA before cell division occur. This process occur during S phase of the cell cycle phases.
DNA replication is the process by which the genetic material is copied prior to distrubution into daughter cells
The original DNA strands are used as templates for the synthesis of new strands
It occurs very quickly, very accurately and at the appropriate time in the life cycle of the cell
General Features of DNA Replication
This process is called semi-conservative because it conserves only half of the original (parent) DNA molecule in the two daughter DNA molecules. One strand in each daughter molecule is completely new.
Complementary Base Pairing.
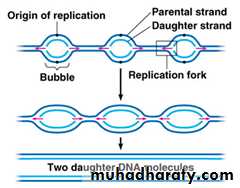
DNA Replication Fork is Asymmetrical.
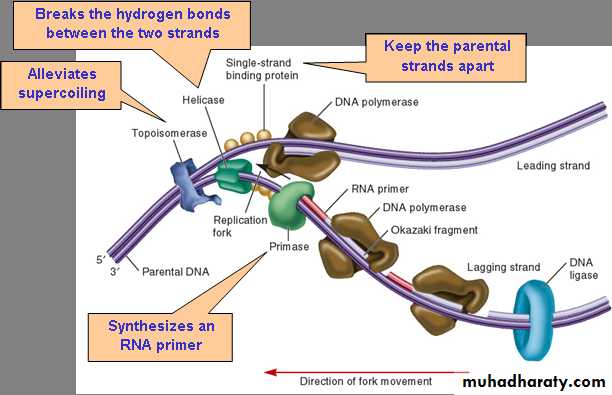
The replication begins with separation of the two DNA strand to form two of parent strand at origin of replication (replication fork). Both strands start together but one of them start by continuous orientation from 3'-5' this strand called leading strand. While the other strand start by discontinuous orientation from 5'-3' this strand called lagging strand, in which the replication occur as a segments by forming an okazaki fragments, primase is used to add a primers at the head of 5' end. In the prokaryotes, there are several enzymes involved in this process:
Helicase separate the double helix of DNA and use of stabilizer to held each newly single strand.
DNA gyrase is used to make sure that the double strand area outside of the replication fork do not supercoil.
DNA polymerase used to catalyzed the addition of new nucleotides to growing daughter strand.
Beta clamp and clamp loader held the DNA polymerase in place of DNA.
Primers a short sequences of RNA, have to be paired to the template strands by the enzyme primase.
DNA polymerase III add short sequences of nucleotides to the primer filling in the gap.
DNA polymerase I used to replace the primers of okazaki fragment by DNA nucleotides.
Ligase is used to ensure bonding between the okazaki fragments.
The process will continue until two identical dsDNA strands are completed by add the telomerase.
Enzyme
Function in DNA replication
DNA Helicase
Also known as helix destabilizing enzyme. Helicase separates the two strands of DNA at the Replication Fork behind the topoisomerase.
DNA Polymerase
The enzyme responsible for catalyzing the addition of nucleotide substrates to DNA in the 5' to 3' direction during DNA replication. Also performs proof-reading and error correction. There exist many different types of DNA Polymerase, (such as the DNA Polymerase III )each of which perform different functions in different types of cells.
DNA clamp
A protein which prevents elongating DNA polymerases from dissociating from the DNA parent strand.
Single-Strand Binding (SSB) Proteins
Bind to ssDNA and prevent the DNA double helix from re-annealing after DNA helicase unwinds it, thus maintaining the strand separation, and facilitating the synthesis of the nascent strand.
Topoisomerase
Relaxes the DNA from its super-coiled nature.
DNA Gyrase
Relieves strain of unwinding by DNA helicase; this is a specific type of topoisomerase
DNA Ligase
Re-anneals the semi-conservative strands and joins Okazaki Fragments of the lagging strand.
Primase
Provides a starting point of RNA (or DNA) for DNA polymerase to begin synthesis of the new DNA strand.
Telomerase
Lengthens telomeric DNA by adding repetitive nucleotide sequences to the ends of eukaryotic chromosomes. This allows germ cells and stem cells to avoid the Hayflick limit on cell division.[23]
Comparison of DNA replication between prokaryotes and eukaryotes
CriteriaProkaryotes
Eukaryotes
Occurrence
Inside the cytoplasm
Inside the nucleus
Replication fork
Only two
Many for each chromosome
Steps
Unwinding of DNA double helix at the origin
HelicaseHelicase
Stabilization of the unbound strand
Single strand binding protein (SSB)
Single strand binding protein (SSB)
Synthesis of DNA strands (leading, lagging strands) (okazaki fragments)
DNA polymerase III
DNA polymerase III
DNA polymerase delta
DNA polymerase alpha
Removal of RNA primer and replacement of RNA to DNADNA polymerase I
Unknown
Joining of Okazaki fragments
DNA ligase I
DNA ligase I
Removal of supercoiled
Topoisomerase II
(DNA gyrase)
Topoisomerase II
(DNA gyrase)
Synthesis of telomers
Not exist
By telomerase
Telomers and Telomerase
Telomerase are distinctive structures found at the ends of our chromosomes. They consist of the same short DNA sequence repeated over and over again. This sequence is usually repeated about 3,000 times and can reach up to 15,000 base pairs? in length. Telomeres serve three major purposes:
They help to organize each of our 46 chromosomes in the nucleus? (control center) of our cells?.
They protect the ends of our chromosomes by forming a cap, much like the plastic tip on shoelaces. If the telomeres were not there, our chromosomes may end up sticking to other chromosomes.
They allow the chromosome to be replicated properly during cell division?:
What happens to telomeres as we age?
Each time a cell divides, 25-200 bases are lost from the ends of the telomeres on each chromosome.
Two main factors contribute to telomere shortening during cell division:
The “end replication problem” during DNA replication: Accounts for the loss of about 20 base pairs? per cell division.
Oxidative stress: Accounts for the loss of between 50-100 base pairs per cell division. The amount of oxidative stress in the body is thought to be affected by lifestyle factors such as diet, smoking and stress.
When the telomere becomes too short, the chromosome reaches a ‘critical length’ and can no longer be replicated.
This ’critical length’ triggers the cell to die by a process called apoptosis?, also known as programmed cell death.
Cell cycle
The sequence of events by which a cell duplicates its genome, synthesizes the other constituents of the cell and eventually divides into two daughter cells is termed cell cycle. Cell division is useful for :Reproduction.
Growth and development.
Renewal.
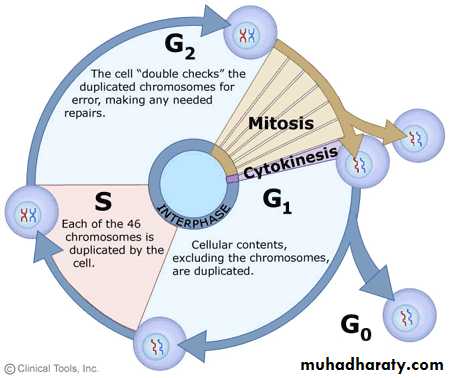
The cell cycle is divided into two basic phases:
Interphase
M Phase (Mitosis phase)
By the end of interphase a cell has two full sets of DNA (chromosomes) and is large enough to begin the division process.
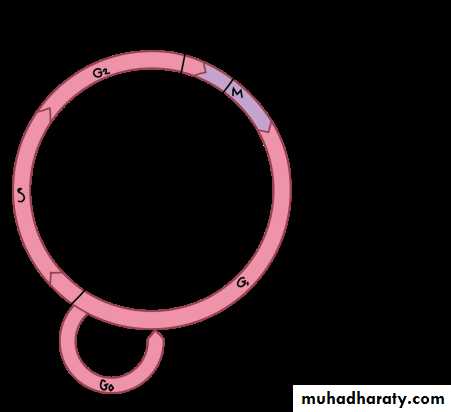
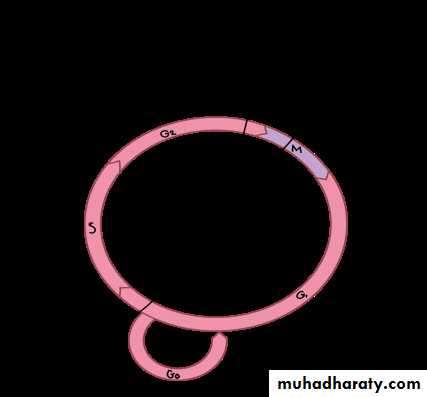
The interphase is divided into three further phases:
G1 phase (Gap 1)
S phase (Synthesis)
G2 phase (Gap 2)
Gap 0 (G0): There are times when a cell will leave the cycle and quit dividing. This may be a temporary resting period or more permanent. An example of the latter is a cell that has reached an end stage of development and will no longer divide (e.g. neuron).
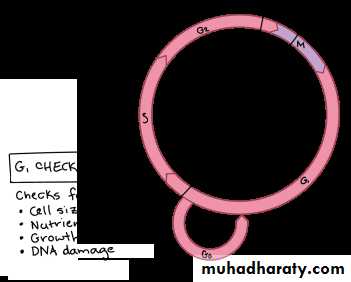
Gap 1 (G1): Cells increase in size in Gap 1, produce RNA and synthesize protein. An important cell cycle control mechanism activated during this period (G1 Checkpoint) ensures that everything is ready for DNA synthesis.
S Phase: To produce two similar daughter cells, the complete DNA instructions in the cell must be duplicated. DNA replication occurs during this S (synthesis) phase.
Gap 2 (G2): During the gap between DNA synthesis and mitosis, the cell will continue to grow and produce new proteins. At the end of this gap is another control checkpoint (G2 Checkpoint) to determine if the cell can now proceed to enter M (mitosis) and divide.
Mitosis or M Phase: Cell growth and protein production stop at this stage in the cell cycle. All of the cell's energy is focused on the complex and orderly division into two similar daughter cells. Mitosis is much shorter than interphase, lasting perhaps only one to two hours. As in both G1 and G2, there is a Checkpoint in the middle of mitosis (Metaphase Checkpoint) that ensures the cell is ready to complete cell division.
Cell cycle checkpoints
A checkpoint is a stage in the eukaryotic cell cycle at which at which the cell examines internal and external cues and "decides" whether or not to move forward with division.There are a number of checkpoints, but the three most important ones are:
Cell cycle regulators (cyclins+ cdk)
Cyclins are among the most important core cell cycle regulators. Cyclins are a group of related proteins, and there are four basic types found in humans and most other eukaryotes: G1 cyclins, G1/S cyclins, S cyclins and M cyclins. Cyclins drive the events of the cell cycle by binding with a family of enzymes called the cyclin-dependent kinases (Cdks) by phosphorylation. A lone Cdk is inactive, but the binding of a cyclins activates it, making it a functional enzyme and allowing it to modify target proteins.
Mitosis-promoting factor (MPF)
Mitotic cyclins accumulate gradually during G2. Once they reach a high enough concentration, they can bind to Cdks. When mitotic cyclins bind to Cdks in G2, the resulting complex is known as Mitosis-promoting factor (MPF). This complex acts as the signal for the G2 cell to enter mitosis. Once the mitotic cyclin degrades, MPF is inactivated and the cell exits mitosis by dividing and re-entering G1.