Diagnostic microbiology
Dr. Ghada YounisDec . 2018
Identifying the organism causing an infectious process is usually essential for effective antimicrobial and supportive therapy.
Initial treatment may be empiric, based on the microbiologic epidemiology of the infection and the patient’s symptoms.
However, definitive microbiologic diagnosis of an infectious disease usually involves one or more of the following five basic laboratory techniques, which guide the physician along a narrowing path of possible causative organisms
: 1) direct microscopic visualization of the organism,
2) cultivation and identification of the organism,3) detection of microbial antigens,
4) detection of microbial DNA or RNA,
and 5) detection of an inflammatory or host immune
response to the microorganism .
PATIENT HISTORY AND PHYSICAL EXAMINATION
A clinical history is the most important part of patient evaluation. Forexample, a history of cough points to the possibility of respiratory tract
infection, whereas dysuria (painful or difficult urination) suggests urinary
tract infection. A history of travel to developing countries may implicate
exotic organisms. For example, a patient who recently swam in the Nile
has an increased risk of schistosomiasis. Patient occupations may suggest exposure to certain pathogens, such as brucellosis in a butcher or anthrax in farmers. Even the age of the patient can sometimes guide the clinician in predicting the identity of pathogens.
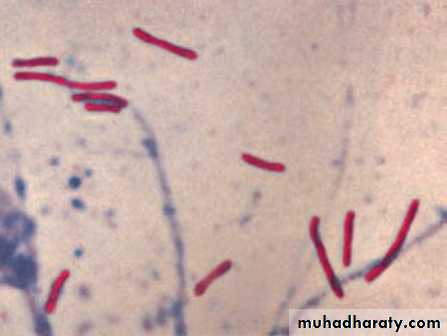
I .DIRECT VISUALIZATION OF THE ORGANISM
In many infectious diseases, pathogenic organisms (excluding viruses)
can often be directly visualized by microscopic examination of patient
specimens, such as sputum, urine, and CSF. The organism’s microscopic
morphology and staining characteristics can provide the first
screening step in arriving at a specific identification. The organisms to
be examined do not need to be alive or able to multiply. Microscopy
yields rapid and inexpensive results and may allow the clinician to initiate treatment without waiting for the results of a culture, as noted in the spinal fluid example in the previous paragraph.
Gram stain
The most common and useful staining procedure is theGram stain, which separates bacteria into two classifications
according to their cell wall composition.
Most, but not all, bacteria are stainable and fall into one of these two groups. [Note: Microorganisms that lack cell walls, such as
mycoplasma, cannot be identified using the Gram stain.]
1. Gram stain applications: The Gram stain is important therapeutically because gram-positive and gram-negative bacteria differ in
their susceptibility to various antibiotics, and the Gram stain may,therefore, be used to guide initial therapy until the microorganism
can be definitively identified. In addition, the morphology of the stained bacteria can sometimes be diagnostic. For example,
gram-negative intracellular diplococci in urethral pus provide ap resumptive diagnosis of gonorrhea.
Acid-fast stain
Stains such as Ziehl-Neelsen (the classic acid-fast stain) are used
to identify organisms that have waxy material (mycolic acids) intheir cell walls. . The most clinically
important acid-fast bacterium is Mycobacterium tuberculosis,
India ink preparation
This is one of the simplest microscopic methods. It is useful indetecting Cryptococcus neoformans in CSF . One drop
of centrifuged CSF is mixed with one drop of India ink on a microscope
slide beneath a glass cover slip. Cryptococci are identified by
their large, transparent capsules that displace the India ink particles.
II .GROWING BACTERIA IN CULTURE
Culturing is routine for most bacterial and fungal infections but is rarelyused to identify helminths or protozoa. Culturing of many pathogens is
straightforward, for example, streaking a throat swab onto a blood agar
plate in search of group A β-hemolytic streptococcus. However, certain
pathogens are very slow growing (for example, M. tuberculosis) or are
cultured only with difficulty (for example, Bartonella henselae).
Microorganisms isolated in culture are identified using such characteristics
as colony size, shape, color, Gram stain, hemolytic reactions on
solid media, odor, and metabolic properties. In addition, pure cultures
provide samples for antimicrobial susceptibility testing . The
success of culturing depends on appropriate collection and transport
techniques and on selection of appropriate culture media, because
some organisms may require special nutrients. Also, some media are
used to suppress the growth of certain organisms in the process of identifying others.
Specimen collection
Many organisms are fragile and must be transported to the laboratory
with minimal delay. For example, gonococci and pneumococci
are very sensitive to heating and drying. Samples must be cultured
promptly, or, if this is not possible, transport media must be used to
extend the viability of the organism to be cultured. When anaerobic
organisms are suspected, the patient’s specimen must be protected
from the toxic effect of oxygen
Two general strategies are used to isolate pathogenic bacteria,
depending on the nature of the clinical sample. The first methoduses enriched media to promote the nonselective growth of any bacteria that may be present.
The second approach employs selective media that only allow growth of specific bacterial species from specimens that normally contain large numbers of bacteria (for example,
stool, genital tract secretions, and sputum). Isolation of a bacterium
is usually performed on solid medium. Liquid medium is used to
grow larger quantities of a culture of bacteria that have already been
isolated as a pure culture.
IDENTIFICATION OF BACTERIA
The most widely used identification scheme involves determining themorphologic and metabolic properties of the unknown bacterium and comparing these with properties of known microorganisms.
It is essential to start identification tests with pure bacterial isolates
grown from a single colony.
Single-enzyme tests
Different bacteria produce varying spectra of enzymes.
For example, some enzymes are necessary for the bacterium’s individual metabolism, and some facilitate the bacterium’s ability to compete with other bacteria or establish an infection.
Tests that measure single bacterial enzymes are simple, rapid, and generally easy to interpret.
They can be performed on organisms already grown in culture
and often provide presumptive identification
1. Catalase test: Catalase-positive organisms rapidly produce bubbles when exposed to a solution containing hydrogen peroxide
The catalase test is key in differentiating between
many gram-positive organisms. For example, staphylococci are
catalase positive, whereas streptococci and enterococci are
catalase negative.
2. Oxidase test: The enzyme cytochrome c oxidase can accept electrons from artificial substrates (such as a phenylenediamine derivative), producing a dark, oxidized product
This test assists in differentiating between groups of gram-negative bacteria. Pseudomonas aeruginosa, for example, is oxidase positive.
3. Urease: The enzyme urease hydrolyzes urea to ammonia and carbon dioxide (NH2CONH2 + H2O → 2NH3 + CO2). The ammonia
produced can be detected with pH indicators that change color in
response to the increased alkalinity . The test helps to identify certain species of Enterobacteriaceae, and Helicobacter pylori.
4. Coagulase test: Coagulase is an enzyme that causes a clot to
form when bacteria are incubated with plasma
The test is used to differentiate Staphylococcus aureus (coagulase
positive) from coagulase-negative staphylococci
III .IMMUNOLOGIC DETECTION OF MICROORGANISMS
In the diagnosis of infectious diseases, immunologic methods take
advantage of the specificity of antigen–antibody binding. For example, known antigens and antibodies are used as diagnostic tools in identifying microorganisms. In addition, serologic detection of a patient’s immune response to infection, or antigenic of a pathogen in a patient’s body fluids, is frequently useful.
Immunologic methods are useful when the infecting microorganism is difficult or impossible to isolate or when a previous infection needs to be documented.
Most methods for determining whether antibodies or antigens
are present in patients’ sera or other body fluids require some type of
immunoassay procedure
A. Detection of microbial antigen with known antiserum
These methods of identification are often rapid and show favorablesensitivity and specificity. However, unlike microbial culturing techniques, these immunologic methods do not permit further characterization of the microorganism, such as determining its antibiotic sensitivity or characteristic metabolic patterns.
1. Quellung reaction: Some bacteria having capsules can be identified
directly in clinical specimens by a reaction that occurs whenthe organisms are treated with serum containing specific antibodies
. The Quellung reaction makes the capsule more refractile and thus more visible, but the capsule does not actually swell. This method can be used for all serotypes of
S. pneumoniae, H. influenzae type b, and Neisseria meningitidis
groups A and C.
2. Slide agglutination test: Some microorganisms, such as
Salmonella and Shigella species, can be identified by agglutination
(clumping) of a suspension of bacterial cells on a microscopic
slide. Agglutination occurs when a specific antibody directed
against the microbial antigen is added to the suspension, causing
cross-linking of the bacteria.
B. Identification of serum antibodies
Detection in a patient’s serum of antibodies that are directed against
microbial antigens provides evidence for a current or past infection
with a specific pathogen. A discussion of the general interpretation
of antibody responses includes the following rules: 1) antibody may
not be detectable early in an infection, 2) the presence of antibodies
in a patient’s serum cannot differentiate between a present and a
prior infection, and 3) a significant rise in antibody titer over a 10 to-
14-day period does distinguish between a present or prior infection.
Techniques such as complement fixation and agglutination can be
used to quantitate antimicrobial antibodies.
1. Complement fixation: One older but still useful method for detecting
serum antibody directed against a specific pathogen employsthe ability of antibody to bind complement .
2. Direct agglutinationDirect bacterial agglutination testing is
sometimes ordered when a suspected pathogen is difficult or
dangerous to culture in the laboratory. This test measures the
ability of a patient's serum antibody to directly agglutinate specific
killed (yet intact) microorganisms. This test is used to evaluate
patients suspected of being infected by Brucella abortus for example
3. Enzyme-linked immunosorbent assay: Enzyme-linked
immunosorbent assay (ELISA)
4. Fluorescent-antibody tests: Organisms in clinical samples can be
detected directly by specific antibodies coupled to a fluorescent
compound such as fluorescein
IV. NUCLEIC ACID –BASED TESTS
The most widely used methods for detecting microbial DNA fall intothree categories:
1) direct hybridization (nonamplified assay),
2) amplification methods using the polymerase chain reaction (PCR)1 or one its variations, and
3) DNA microarrays. Although not likely to completely replace culture techniques in the near future, nucleic acid–based tests for the diagnosis of infectious diseases are gaining wider acceptance as more products approved by the Food and Drug Administration become commercially available.
Advantages of polymerase chain reaction:
Methods employing nucleic acid–amplification techniques have a major advantage over direct detection with nucleic acid probes because amplification methods allow specific DNA or RNA target sequences of the pathogen to be amplified millions of times without having to culture the microorganism itself for extended periods.PCR also permits identification of noncultivatable or slow-growing microorganisms, such as mycobacteria, anaerobic bacteria, and
viruses.
Nucleic acid–amplification methods are sensitive, specific for the target organism, and are unaffected by the prior administration of antibiotics
. Applications: Nucleic acid–amplification techniques are generally
quick, easy, and accurate. A major use of these techniquesis for the detection of organisms that cannot be grown in vitro or
for which current culture techniques are insensitive. Moreover,
they are useful in the detection of organisms that require complex
media or cell cultures and/or prolonged incubation times
Limitations: PCR amplification is limited by the occurrence of
false-positives due to cross-contamination with other
microorganisms’ nucleic acid. PCR tests are often costly and
require skilled personnel.
V. SUSCEPTIBILITY TESTING
After a pathogen is cultured, its sensitivity to specific antibiotics serves as a guide in choosing antimicrobial therapy. Some pathogens, such as Streptococcus pyogenes and N. meningitidis, usually have predictable sensitivity patterns to certain antibiotics. In contrast, most gram-negative bacilli, enterococci, and staphylococcal species show unpredictable sensitivity patterns to various antibiotics and require susceptibility testing to determine appropriate antimicrobial therapy.A. Disk-diffusion method
The classic qualitative method to test susceptibility to antibiotics hasbeen the Kirby-Bauer disk-diffusion method, in which disks with
exact amounts of different antimicrobial agents are placed on culture
dishes inoculated with the microorganism to be tested. The
organism’s growth (resistance to the drug) or lack of growth (sensitivity
to the drug) is then monitored . In addition, the size of the zone of growth inhibition is influenced by the concentration and rate of diffusion of the antibiotic on the disk. The disk - diffusion method is useful when susceptibility to an unusual antibiotic, not available in automated systems, is to be determined
B. Minimal inhibitory concentration
Quantitative testing uses a dilution technique in which tubes containingserial dilutions of an antibiotic are inoculated with the organism
whose sensitivity to that antibiotic is to be tested. The tubes are
incubated and later observed to determine the minimal inhibitory
concentration (MIC) of the antibiotic necessary to prevent bacterial
Growth
Quantitative susceptibility testing may be necessary for
patients who either fail to respond to antimicrobial therapy . In some clinical cases, the minimal bactericidal concentration may need to be determined. This is the lowest concentration of antibiotic that kills 100 percent of the bacteria,
rather than simply inhibiting growth, MBC.
Good luck