Course: Virology
Lecturer: Dr. Weam SaadLecture: Viral Taxonomy and Life Cycle
Viral Taxonomy
Viruses classification is dynamic because of new viruses are continuously discovered and more new information known. The classification depends on the International Committee on the Taxonomy of Viruses (ICTV).The basic viral hierarchical classification is:
Order - Family - Subfamily - Genus - Species - Strain / Type.
For example: DNA and RNA Reverse Transcribing Viruses, Hepadnaviridae, Icosahedral, Orthohepadna virus, Hepatitis B virus
The basic viral characteristic is nucleic acid type (DNA or RNA) and morphology, the virion size, shape, and the presence or absence of an envelope. The host type and immunological properties (serotypes) of the virus are also used. Physical and physicochemical properties such as molecular mass, density, thermal inactivation, pH stability, and sensitivity to various solvents are used in classification. Whether the RNA or DNA is single or double stranded, the organization of the genome and the presence of particular genes comprise important aspects of the current taxonomy of viruses.
The basic characters that are used for classification are:
Particle morphology: the shape and size of particles as seen under the electron microscope.
Genome properties: this includes the number of genome components and the translation strategy. Where genome sequences have been determined.
Biological properties: this may include the type of host cell and the mode of transmission.
Serological properties: the relation of the virion proteins.
Also, classification is based upon macromolecules produced (like the structural proteins and enzymes), antigenic properties and biological properties (e.g., accumulation of virions in cells, infectivity, hemagglutination).
Based on the genome; the viruses' classification includes six major groups:
Double-stranded DNA (dsDNA): there are no plant viruses in this group, which includes only the viruses that replicate without an RNA intermediate. It includes those viruses with the largest known genomes (up to about 400,000 base pairs) and there is only one genome component, which may be linear or circular. Examples; the herpes and pox viruses.
Single-stranded DNA (ssDNA): there are two families of plant viruses in this group and both of these have small circular genome components, often with two or more segments.
Reverse-transcribing viruses: these have dsDNA or ssRNA genomes and their replication includes the synthesis of DNA from RNA by the enzyme reverse transcriptase; many integrate into their host genomes. The group includes the retroviruses (Human immunodeficiency virus (HIV), the cause of AIDS). There is a single family of plant viruses in this group and have a single component of circular dsDNA, the replication is via an RNA intermediate.
Double-stranded RNA (dsRNA): some plant viruses and many of the mycoviruses are included in this group.
Negative sense single-stranded RNA (ssRNA-): in this group, some or all of the genes are translated into protein from an RNA strand complementary to that of the genome (as packaged in the virus particle). There are some plant viruses in this group and it also includes the viruses that cause measles, influenza and rabies.
Positive sense single-stranded RNA (ssRNA+): the majority of plant viruses are included in this group. It also includes the SARS coronavirus and many other viruses that cause respiratory diseases (including the "common cold"), and the viruses of polio and foot-and-mouth disease.
According to the shapes they are classified as followings:
Isometric: apparently spherical and (depending on the species) from about 18 nm in diameter and more. The example in the picture shows Tobacco necrosis virus, genus Necrovirus.
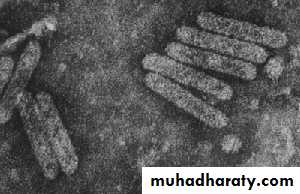
Rod-shaped: about 20-25 nm in diameter and from about 100 to 300 nm long. These appear rigid and often have a clear central canal (depending on the staining method used). Some viruses have two or more different lengths of particle and these contain different genome components. The example in the picture shows Tobacco mosaic virus.
Filamentous: usually about 12 nm in diameter and more flexible than the rod-shaped particles. They can be up to 1000 nm long, or even longer. Some viruses have two or more different lengths of particle and these contain different genome components. The example in the picture shows Potato virus Y, genus Potyvirus.
Geminate: twinned isometric particles about 30 x 18 nm. These particles are diagnostic for viruses in the family Geminiviridae which are widespread in many crops especially in tropical regions. The example in the picture shows Maize streak virus, genus Mastrevirus.
Bacilliform: Short round-ended rods. These come in various forms up to about 30 nm wide and 300 nm long. The example here shows Cocoa swollen shoot virus, genus Badnavirus.
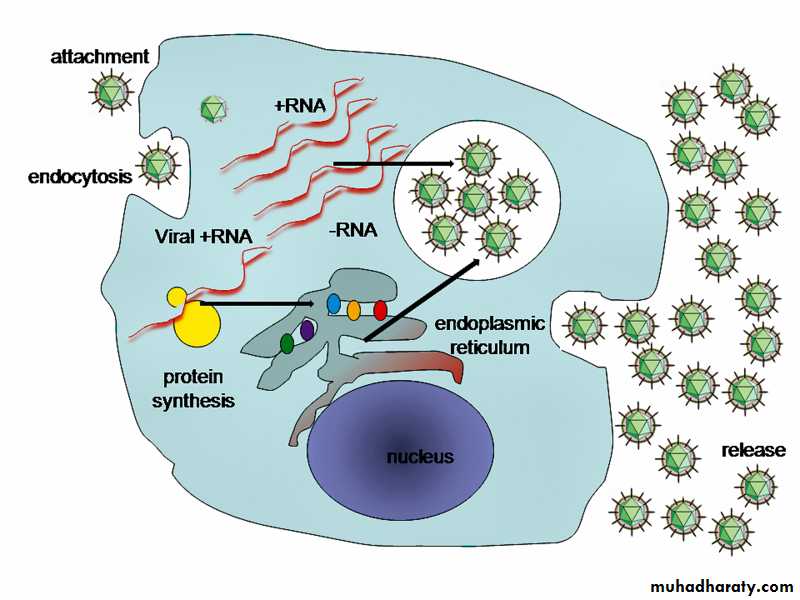
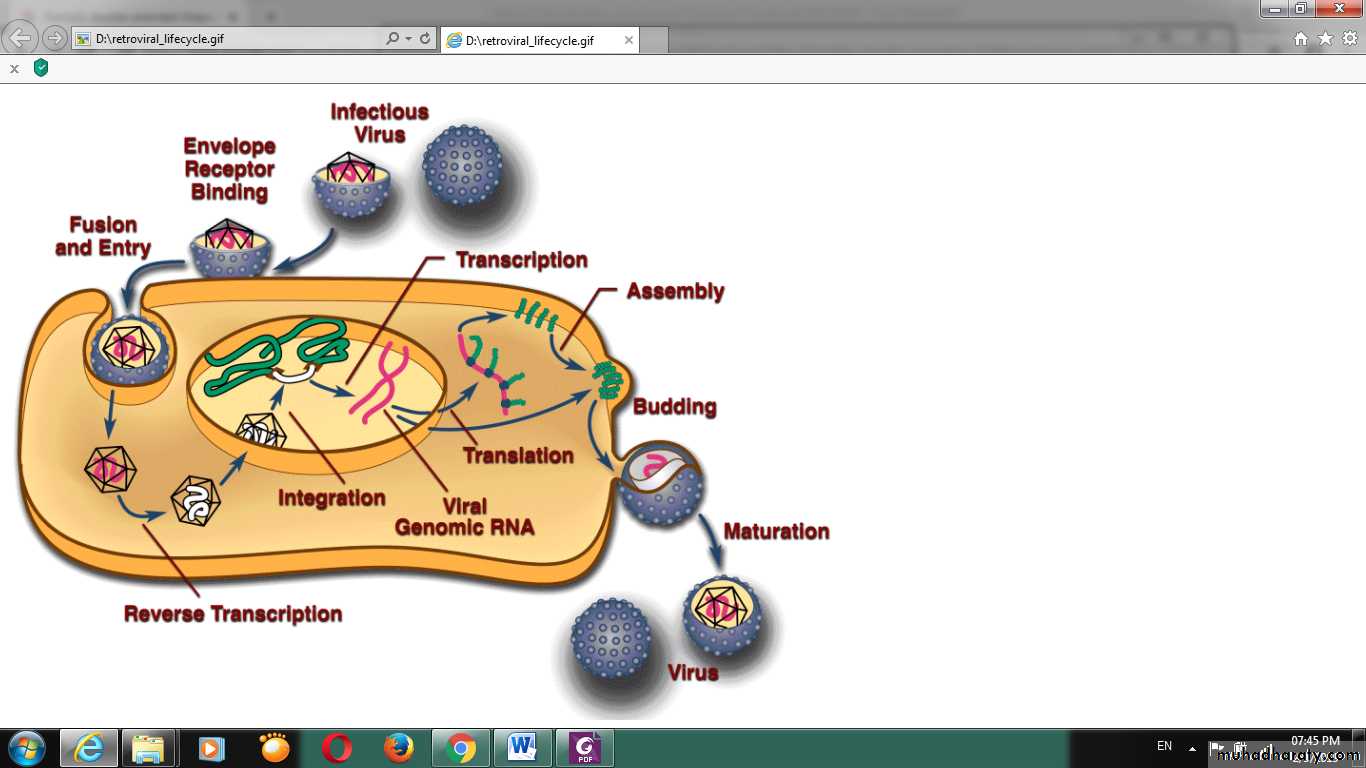
Replication of virus (life cycle):
Viruses multiply only in living cells because they are obligate intracellular parasites. The host cells give the energy and synthetic systems or machinery for the synthesis of viral proteins and nucleic acids. The viral nucleic acid carries the necessary genetic codes for all of the virus macromolecules.Attachment
First stage of virus life cycle starts by a specific binding of viral capsid proteins to the specific receptors on the host cellular surface. Each virus has specific host cell and that was used in classification of viruses, e.g., HIV infects the human lymphocytes T-cells. This is because its surface protein, gp120, specifically interacts with the CD4 molecule (a chemokine receptor) which is most commonly found on the surface of CD4+ T-Cells, poliovirus is able to attach only to the central nervous system and intestinal tract cells of human and animals. Attachments to the receptor induce the viral envelope protein to change and cause the fusion of virus and cellular membranes, or changes of non-enveloped virus surface proteins that allow the virus to enter inside host cell.Penetration :
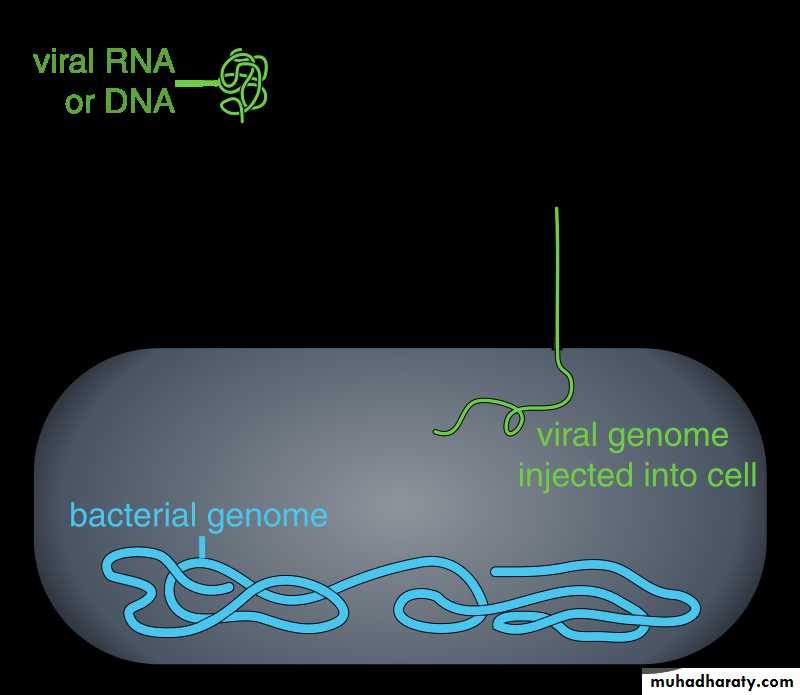
Virions are another name for a virus particle, an entire virus particle during transmission of the nucleic acid genome from one host cell to another host cell. The virions enter the host cell through receptor mediated endocytosis, engulfment or membrane fusion. This is called viral entry.The infection of plant and fungal cells is different from that of animal cells. Plants have a rigid cell wall made of cellulose, and fungi cell wall made of chitin, so most specific viruses can get inside these cells only after trauma of the cell wall, all plant viruses (such as tobacco mosaic virus) can also move directly from cell to cell, in the form of single-stranded nucleoprotein complexes, through pores called HYPERLINK "https://en.wikipedia.org/wiki/Plasmodesma" \o "Plasmodesma" plasmodesmata. Also, the bacteria have strong cell walls against viral penetration, but some viruses (bacteriophages) developed mechanisms that inject their genome into the bacterial cell across the cell wall and the viral capsid remains outside.
Uncoating:
This stage includes the remove of viral capsid by degradation using viral enzymes or host enzymes or by simple dissociation; the end-result is the releasing of the viral genomic nucleic acid inside host cell cytoplasm. The infectivity of the parental virus is lost at the uncoating stage.Replication:
This stage involves multiplication of the genome. Replication involves synthesis of viral messenger RNA (mRNA) transcribed from the viral nucleic acid.DNA viruses replicate in the nucleus and use host cell DNA and RNA polymerases and processing enzymes in their replication. But RNA viruses genome is replicated in host cell cytoplasm. Viral protein and other components synthesis occur after translation the mRNA, and then viral genome replication mediated by regulatory protein expression. In complex viruses, this stage needs further rounds of mRNA synthesis and gene expression for structural proteins. Some viruses (eg, rhabdoviruses) carry RNA polymerases to synthesize mRNAs.
DNA viruses replication:
The genome replication occurs in the nucleus, these viruses can enter the host cell by direct fusion with the cell membrane (e.g., herpesviruses) or by specific receptor-mediated endocytosis. In the eukaryotic host cells the viral genome must cross the cell's nuclear membrane to access this machinery or system for DNA replication, while in bacteria they need only to enter the host cell. In double-stranded DNA viruses, viral proteins are synthesized soon after infection when viral DNA synthesis begins.
RNA viruses replication:
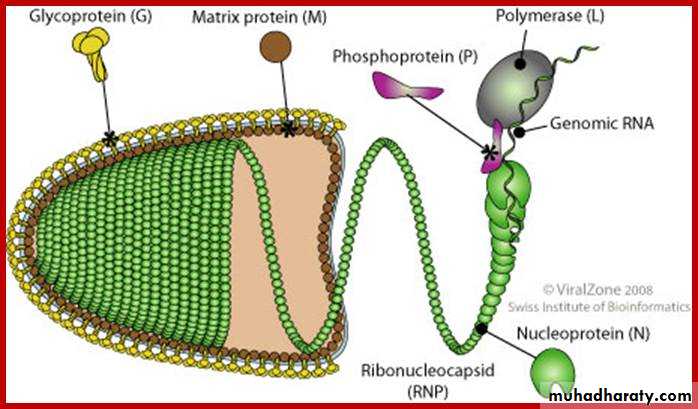
They all use their own RNA replication enzymes (replicase) to create copies of their genomes and replication occurs in the cytoplasm of host cell. The ssRNA viruses are called negative-strand (negative sense viruses) because their single-strand RNA genome is complementary to mRNA, which is conventionally designated positive strand (positive sense). The negative strand viruses have their own RNA polymerase because eukaryotic cells lack enzymes able to synthesize mRNA of an RNA template. Some virions carry polymerases e.g. (orthomyxoviruses, reoviruses).
Both types ssRNA viruses e.g. Retroviridae and dsDNA viruses e.g. Hepatitis B virus use a reverse transcriptase, or RNA-dependent DNA polymerase enzyme, to carry out the nucleic acid conversion
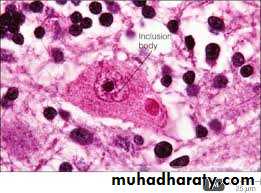
During viral multiplication within host cells, virus specific structures called Inclusion bodies sometimes called elementary bodies may be produced. They are larger than the virus particle contains host protein, ribosomal components or DNA/RNA fragments and can be stained by dyes (eg, eosin or acid dye). They may be formed in the nucleus (e.g. herpesvirus), in the cytoplasm (e.g. poxvirus), or in both nucleus and cytoplasm (e.g. measles virus). In many viral infections, the inclusion bodies are the site of development for the virions (the viral factories). The inclusion bodies are diagnostic for the infection with the virus. The intra cytoplasmic inclusion in nerve cells (the Negri body) is pathognomonic for rabies viral infection.
(Inclusion body after viral infection)
Inclusion bodies sites and examples:Intra-cytoplasmic:
Negri bodies in Rabies
Guarnieri bodies in HYPERLINK "https://en.wikipedia.org/wiki/Vaccinia" \o "Vaccinia" vaccinia, HYPERLINK "https://en.wikipedia.org/wiki/Variola" \o "Variola" variola (smallpox)
Paschen bodies in HYPERLINK "https://en.wikipedia.org/wiki/Variola" \o "Variola" variola (smallpox)
Intra-nuclear:
Cowdry type A in Herpes simplex virus and Varicella zoster virus
Torres bodies in Yellow fever
Cowdry type B in Polio and adenovirus
"Owl's eye appearance" in cytomegalovirus
Both Intra-cytoplasmic and Intra-nuclear: e.g. Warthin–Finkeldey bodies in Measles.
Assembly:
This step includes self-assembly of the virus particles, to form the virus structure, synthesized viral genomes and capsid polypeptides assemble together to form progeny viruses, some modification of the proteins occurs usually called maturation. In viruses such as HIV, this modification occurs after the virus release from the host cell.
The non-enveloped viruses accumulate in infected cells, and the cells eventually lyse and release the virus particles. While, enveloped viruses mature by a budding process. Virus glycoproteins specific for the envelope are inserted into cellular membranes; viral nucleocapsids then bud through the membrane to form an envelope. Budding then occurs at the plasma membrane (sometimes involve other membranes in the host cell). Enveloped virusesare not infectious until they have acquired their envelops.
Excess amounts of viral components may accumulate and be involved in the formation of inclusion bodies in the cell. The cellular cytopathic effects develop and the host cells die. But, sometimes the host cell is not damagedby the virus (long-term persistent infections) e.g. herpes virus. Some viruses induce mechanisms to regulate apoptosis in host cell and cause host cells self-destruction. Some viruses, such as Epstein–Barr virus, can cause proliferation of host cells without causing malignancy, while other viruses such as papilloma viruses, are the causes of cancer; these are called oncogenic viruses because they stimulate uncontrolled cell growth, causing the transformation or immortalization of the cell.
Release:
The virus is released from the cell through exocytosis or through budding (e.g. many enveloped viruses) from the plasma membrane; other Viruses can be released from the host cell by HYPERLINK "https://en.wikipedia.org/wiki/Lysis" \o "Lysis" lysis of host cell (a process that kills the cell by bursting the membrane and cell wall if present, e.g. many bacterial and some animal viruses). This is called a lysogenic cycle where the viral genome is incorporated (linked) by genetic recombination into a specific place in the host's chromosome. The viral genome is then known as a "provirus" or, in the case of bacteriophages a " HYPERLINK "https://en.wikipedia.org/wiki/Prophage" \o "Prophage" prophage".Enveloped viruses (e.g., HIV) typically are released from the host cell by budding. During this process the viruses get the envelope, which is a modified piece of the host's plasma membrane or other internal membranes of the host cells as mentioned before.
(RNA viruses' life cycle inside host cell)
(DNA viruses' life cycle inside host cell)