The four orders of protein structure
primary structure :- the primary structure of the polypeptide chain of a
protein is the order in which amino acids are joined together, and it
includes the location of any disulfide bonds.
The number of known protein sequences is so large and is increasing so
rapidly that rather than being available in printed from, sequence data
are now deposited in electronic protein sequence databases that can
be accessed via that internet .
secondary structure :- "
Configuration and conformation
"
The term configuration refers to the geometric relationship between a
given set of atoms . Interconversion of configurationally alternatives.
(eg, conversion of D- to L- alanine ) can be achieved only by breaking
and re-forming covalent bonds.
The similar - sounding term "conformation" refers to the Three-
dimensional architecture of a protein, the spatial relationship of all the
atoms to all the others .
The interconversion of conformers involves not the rupture of covalent
bonds but the rupture and reformation of noncovalent forces (
hydrogen bonds, salt bonds, hydrophobic interaction ) that stabilize
given conformations. Even after ruling out conformations precluded by
steric
interactions, the free rotation about two-thirds of the covalent
bonds of the main chain of a polypeptide allows for a staggeringly large
number of possible conformations for a given protein however, for a
given protein, only a small number of possible conformations have
biologic significance
Various forces stabilize protein structures
several individually weak but numerically formidable noncovalent
interaction stabilize protein conformation.
These forces include hydrogen bonds, hydrophobic interaction,
electrostatic interaction, and van der waals forces.
HYDROGEN BONDS: Residues with polar R groups generally on the
surface of globular protein, where they form hydrogen bonds primarily
to water molecules. Elsewhere the aminoacyl residues of the backbone
form hydrogen bonds with one another.
HYDROPHOBIC INTERACTIONS: Hydrophobic interactions involve the
nonpolar R groups of aminoacyl residues that in typical globular protein
reside in the interior of the protein. Formation of hydrophobic
interaction is "entropically driven" . A roughly spherical overall shape
minimizes surface area. Concentration of nonpolar residues in the
interior of the protein lowers the number of surface residues and
maximizes the opportunity for the film of surface water molecules to
form hydrogen bonds with one another, a process associated with an
increase in entropy. By contrast, the nonpolar environment of
biological membranes favors hydrophobic surface residues whose
nonpolar R groups participate in hydrophobic interaction with the alkyl
side chain of the fatty acyl esters of membrane bilayers ..
Electrostatic interaction : or salt bonds are formed between oppositely
charged groups such as the amino terminal and carboxyl terminal
groups of peptides and the charged R groups of polar aminoacyl
residues , while all formally charged groups tend to be located on the
surface of globular proteins, exceptions occur. specific polar groups
that perform essential biologic functions may reside in clefts that
penetrate the interior of a protein . since polar residues can also
participate in ionic interactions the presence of salts such as KCL can
significantly decrease ionic interactions between surface residues.
Van der waals interactions : Van der waals forces which are extremely
weak and act only over extremely short distances, include both an
attractive and a repulsive component. The attractive force involves
interaction between induced
dipole
formed by momentary fluctuations
in the electron distribution in nearby atom. The repulsive force comes
into play when two atom come so close that their electron orbitals
overlap. The distance of which the attractive force is maximal and the
repulsive force is minimal is termed the van der waals radii. Atoms have
characteristics van der waals radii and the optimal contact distance
between two atom is the sum of their van der waals radii..
Tertiary structure : Fig 6-10 P54
The high information content of diagrams or models that include all
atoms of a protein hinders the study of overall features of protein
structure. Consequently, we employ simplified conventional
representation to display structure featurs. The symbols used are
cylinders for α-helices. broad arrows for
β- strands, and ribbon like
strands for the remaining structures such as β-bends and loops
..
Electrostatic bonds like surface residues :-
Salt (electrostatic) bonds link oppositely charged R groups of residues
and the charged α- groups of carboxyl and amino terminal residues. For
example , the R group of Lysine ( Net charged +1 at physiologic PH )
and aspartate or glutamate ( Net charged -1 ) can interact
Electrostatically to stabilize protein .
Disulfide bonds confer additional stability :
In addition to peptide bonds, covalent Disulfide bonds can form
between Cysteine residues present in the same or different
polypeptide. these disulfide bonds confer additional stability to specific
conformation of proteins such as enzyme ( eg: Ribonuclease ) and
structural proteins ( eg; KERATIN ) .
hydrophobic interaction link interior residual :
Nonpolar side chains of amino acids associate in the interior of globular
protein. while these associations are individually extremely weak, these
large number
dictate
that hydrophobic interaction contribute
significantly to maintaining protein structure …
QUATERNARY STRUCTURE
oligomeric protein have multiple polypeptide chains !
protein that contain two or more polypeptide chain associated by
noncovalent forces are said to exhibit quaternary structure in these
multimeric protein the individed polypeptide chains are termed
protomers or subunits .
Hydrogen bonds and electrostatic bonds formed between surface
residues of adjacent subunits stabilize the association of subunits .
protein composed of two or four subunits are termed dimeric or
tetrameric proteins, respectively, homodimers, homotetramers, etc.
consist of identical subunits, hetero-oligomers of dissimilar subunits.
The different subunits of hetero-oligomers protein typically perform
discrete functions. one subunit or set of identical subunits may perform
a catalytic function , another subunit set, ligand recognition or a
regulatory role. Different spatial orientations of subunit confer
alterative properties on the oligomer and pemit multimeric proteins to
play unique roles in intercellular regulation..
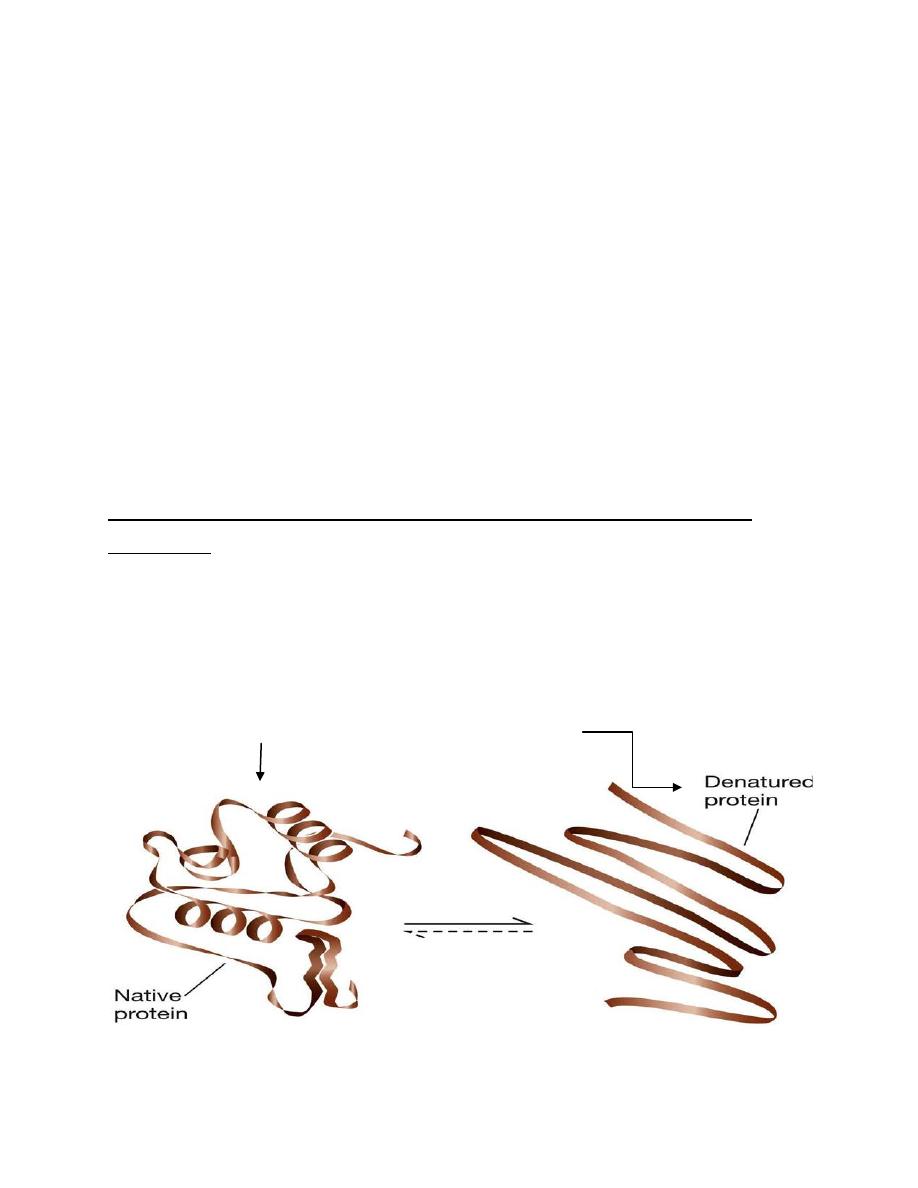
Protein denaturant, disrupt secondary , tertiary , and quaternary
structure .
Reagents' such as urea, sodium dodecyl sulfate (SDS), mild H+ and mild
OH- rupture hydrogen bonds, hydrophobic bonds, and electrostatic
bonds ( but not peptide or disulfide bonds). They thus disrupt all the
orders of protein structure except PRIMARY STURUCTE and destroy
biologic activity . (fig)
active
------------------------ inactive
representation of denaturation of a protomer

physical methods reveal molecular weight and Quaternary structure
determination of the quaternary structure of oligomeric proteins
involves determining the number and kind of promoters present, their
mutual orientation and the interactions that unite them . providing that
oligomers do NOT undergo denataration during the procedure
used to determine molecular weight, many methods can yield
molecular weight data.
These same techniques may be used to determine protomer molecular
weight if the oligomer is first dentaturated .
A- analytical ultracentrifugation .
B- sucrose density gradient centrifugation.
C- Gel filtration .
D- polyacrylamide gel electrophoresis (PAGE ).
E- Electron microscopy visualizes macromolecular complexes .
By Oday D. Abdulqader ..
اﻟﺳﻼم ﻋﻠﯾﻛم ھذه اﻟﻣﻼﺣظﻼت واﻟﺻور ﻟﻼطﻼع ﻓﻘط واﻟﻔﮭم ﻣﺎ اﻟﮭﺎ ﻋﻼﻗﮫ ﺑﻣﺎدة
اﻟدﻛﺗورة اﺗﻣﻧﺎﻟﻛم اﻟﺗوﻓﯾﻖ
..
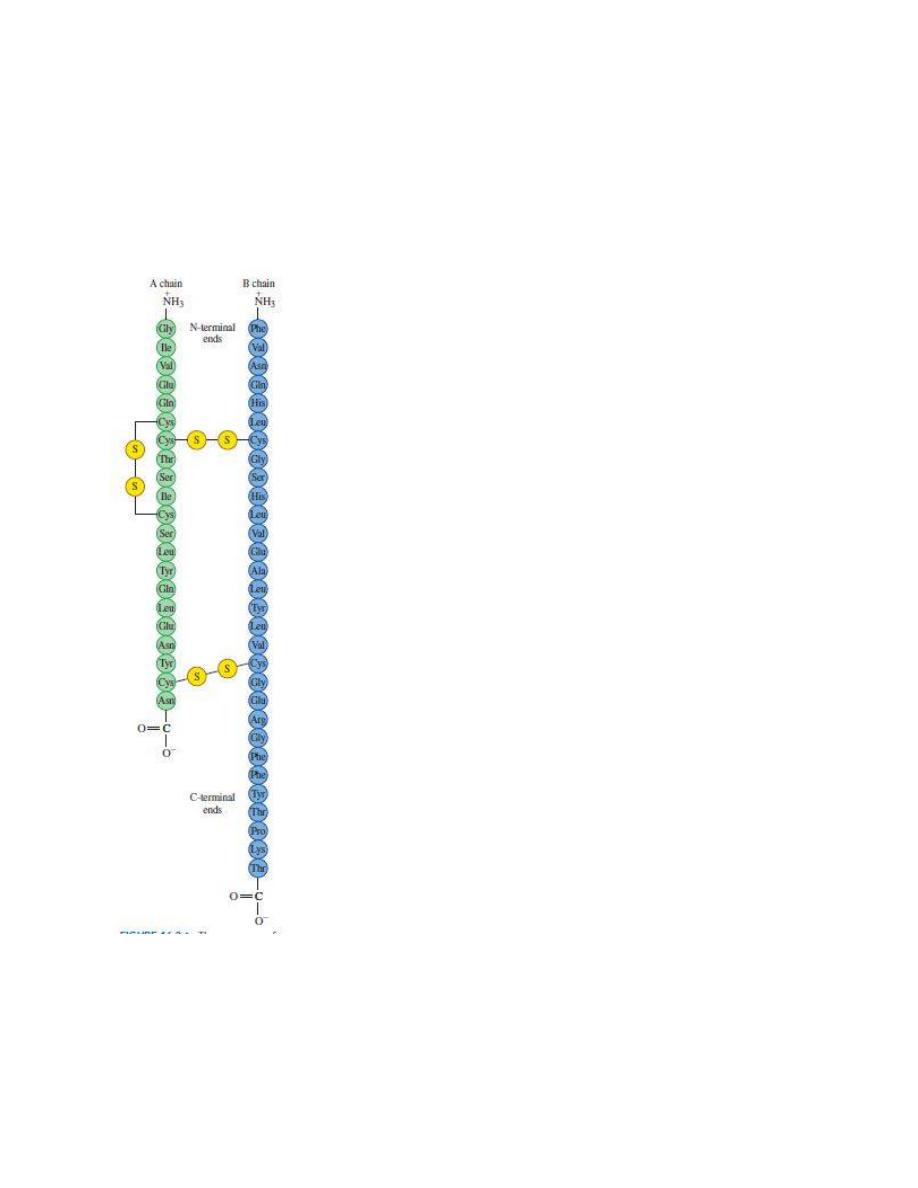
The sequence of amino acids in human insulin is its primary structure.
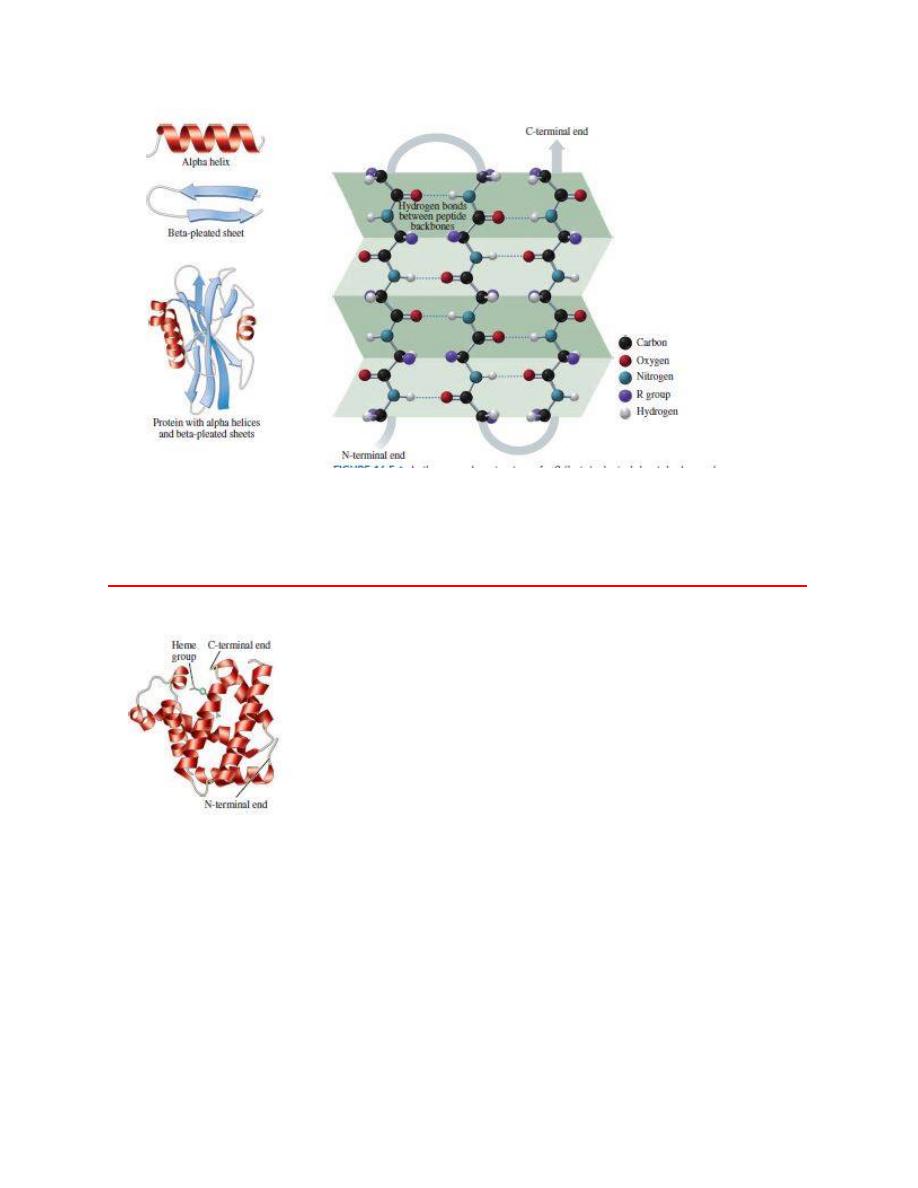
In the secondary structure of
α
b 1beta2-pleated sheet, hydrogen bonds
form between the peptide chains.
---------------------------------------------------------------------------------------------------------
The ribbon model represents the tertiary structure of the polypeptide chain of myoglobin, which
is a globular protein that contains a heme group that binds oxygen.
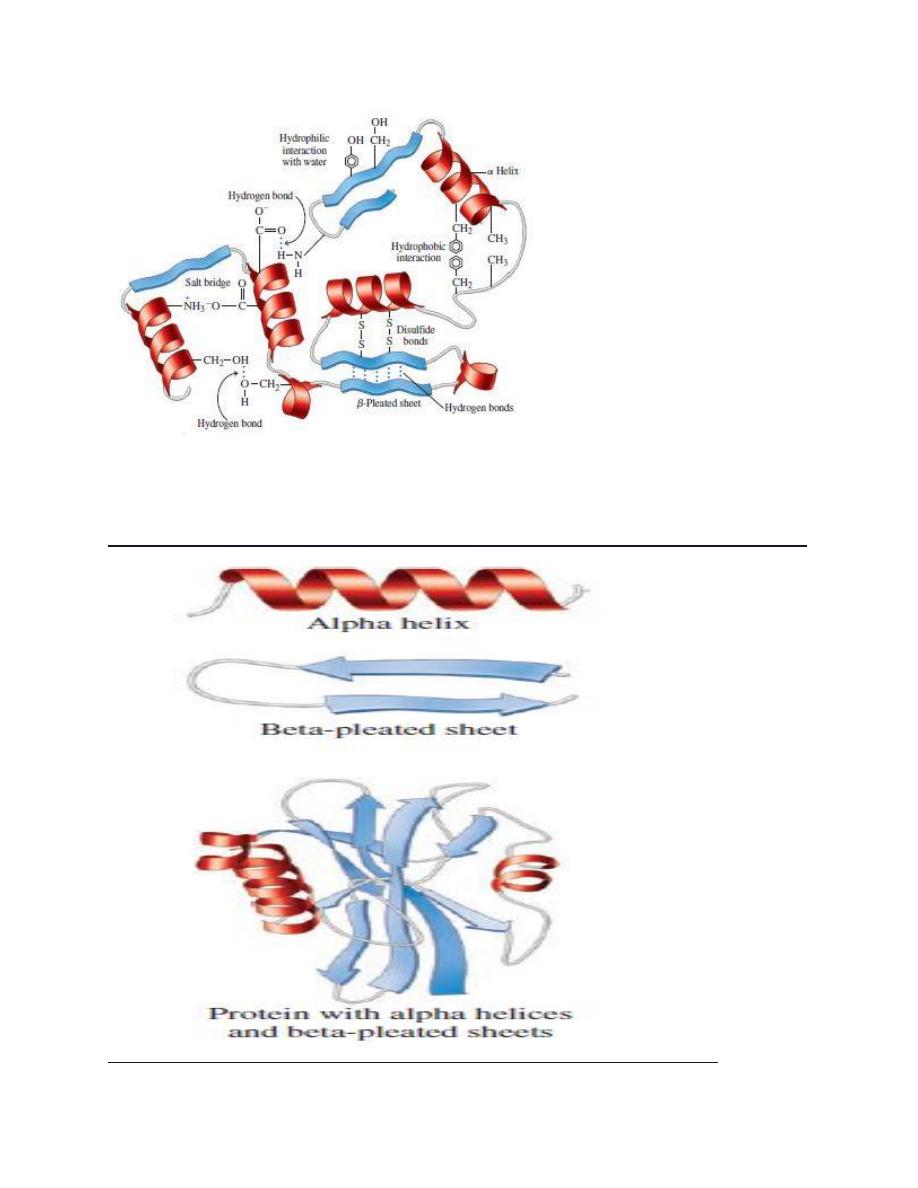
Interactions between amino acid R groups fold a protein into a specific
three-dimensional shape called its tertiary structure.
=========================================================
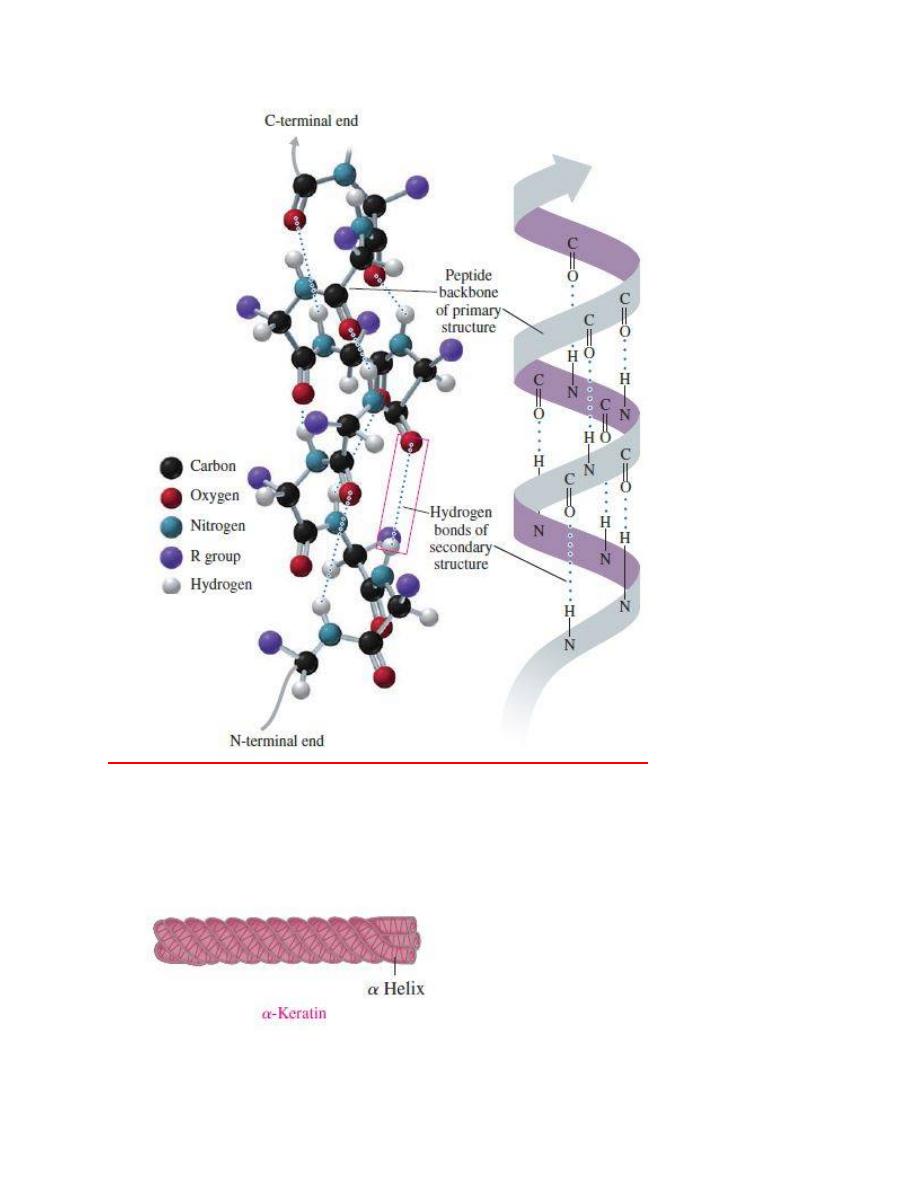
The
α helix acquires a coiled shape from hydrogen bonds between the oxygen of the
C
═O group and the hydrogen of the N¬H group in the next turn.
----------------------------------------------------------------------------------------------------
The fibrous proteins of
α-keratin wrap together to form fibrils of hair and wool.
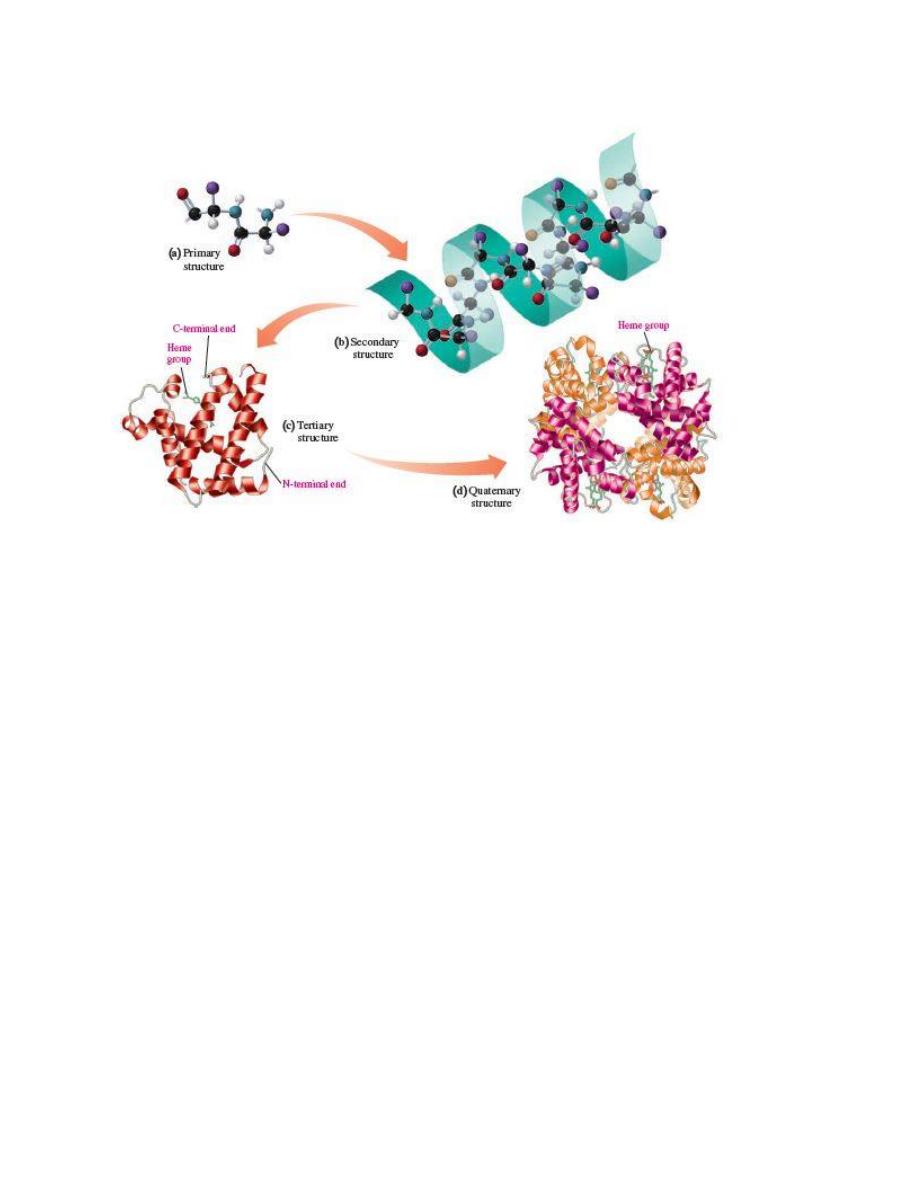
The structural levels of protein are(a) primary, (b) secondary, (c)
tertiary, and sometimes (d) quaternary
