
Lecture 6: Protein targeting and degradation
Prof. Dr. Hedef Dhafir El-Yassin 2013
1
Protein Targeting
Objectives
1. to understand how proteins find their destination in
prokaryotic and eukaryotic cells
2. to know how proteins are bio-recycled
As a protein is being synthesized, decisions must be taken about
sending it to the correct location in the cell where it will be
required. The information for doing this resides in the nascent
protein sequence itself. Once the protein has reached its final
destination, this information may be removed by proteolytic
processing.
Targeting in Bacteria
In bacterial cells, the targeting decision is relatively straightforward:
is the protein destined to be an
intracellular
protein or an
extracellular
one?
Lecture 6: Protein targeting and degradation
Prof. Dr. Hedef Dhafir El-Yassin 2013
2
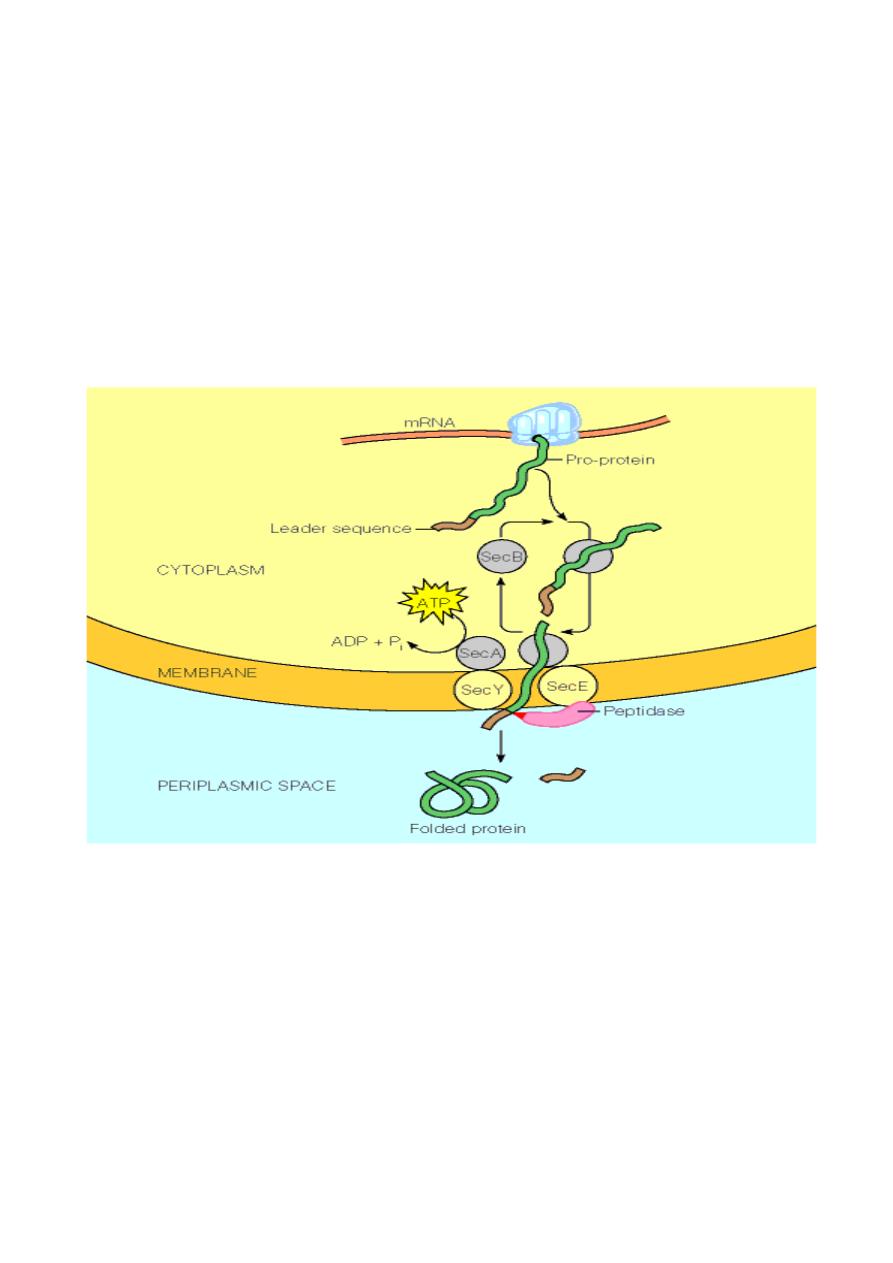
Secreted proteins contain a
signal sequence
. This is a short (6 -
30) stretch of hydrophobic amino acids, flanked on the N-terminal
side by one or more positively charged amino acids such as lysine
or arginine, and containing neutral amino acids with short side-
chains (such as glycine or alanine) at the cleavage site. As
proteins with signal sequences are synthesized, they are bound by
the
SecB
protein. This prevents the protein from folding.
SecB
delivers the protein to the cell membrane where is secreted
through a pore formed by the
SecE
and
SecY
proteins. Secretion
is driven by the
SecA
ATPase. After the protein has been
secreted, the signal sequence is removed by a membrane bound
leader peptidase
.
Lecture 6: Protein targeting and degradation
Prof. Dr. Hedef Dhafir El-Yassin 2013
3
Targeting in Eukaryotes
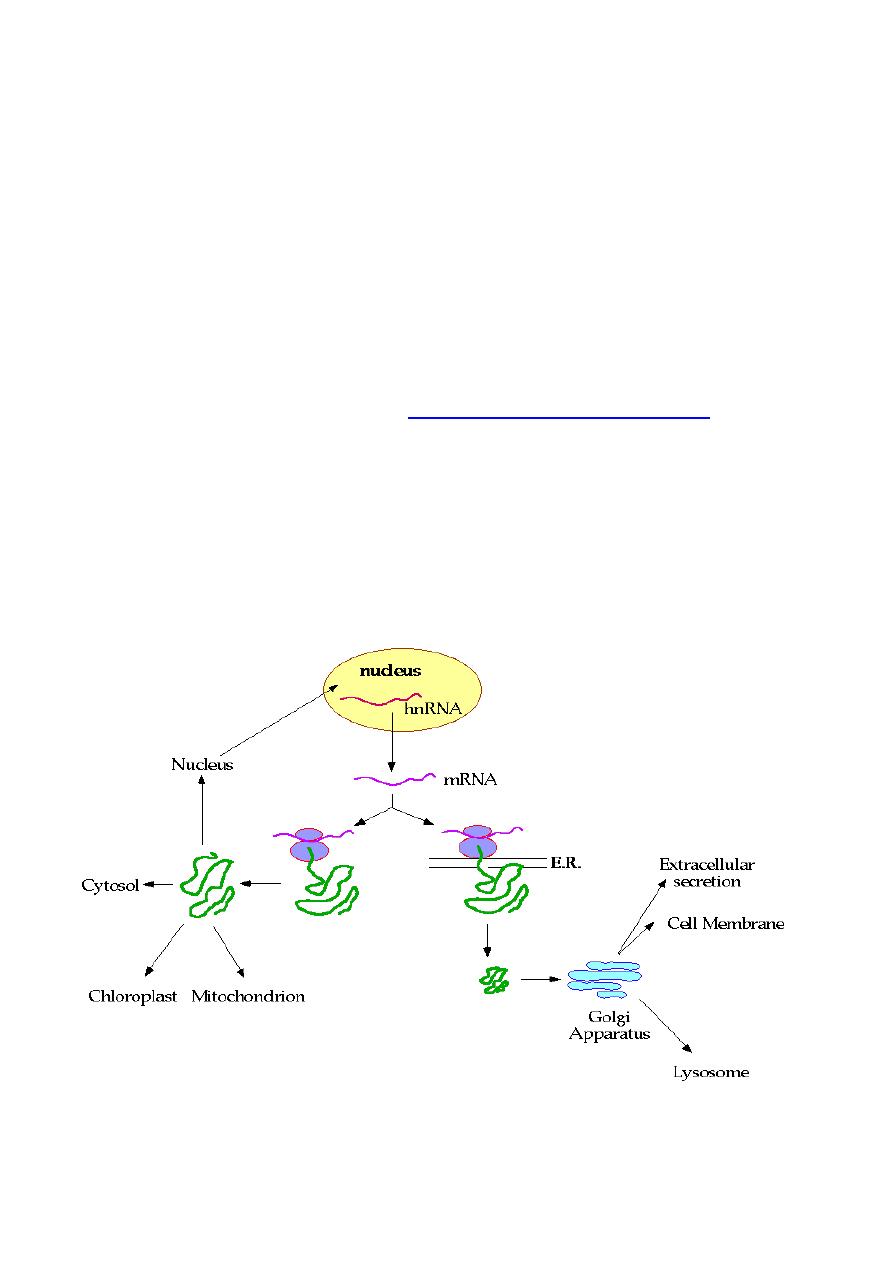
In eukaryotic cells, the situation is more complex.
Extracellular
proteins
can be targeted for
secretion
or to the
cell membrane
,
or to one of the many
internal organelles
such as the lysosome.
Intracellular proteins
can be targeted for the
cytoplasm
, to the
nucleus
or to special
organelles
such as the mitochondrion.
The Signal Sequence hypothesis was first enunciated by Gunther
Blöbel who was awarded the
Nobel Prize in Medicine in 1999
his work.
The following diagram summarizes the choices/fates available to
newly synthesized proteins in a eukaryotic cell:
Lecture 6: Protein targeting and degradation
Prof. Dr. Hedef Dhafir El-Yassin 2013
4
Protein secretion in eukaryotic cells also involves a signal
sequence. As in bacteria, the signal sequence is a short (6 - 30)
stretch of hydrophobic amino acids, flanked on the N-terminal side
by one or more positively charged amino acids such as lysine or
arginine, and containing neutral amino acids with short side-chains
(such as glycine or alanine) at the cleavage site:
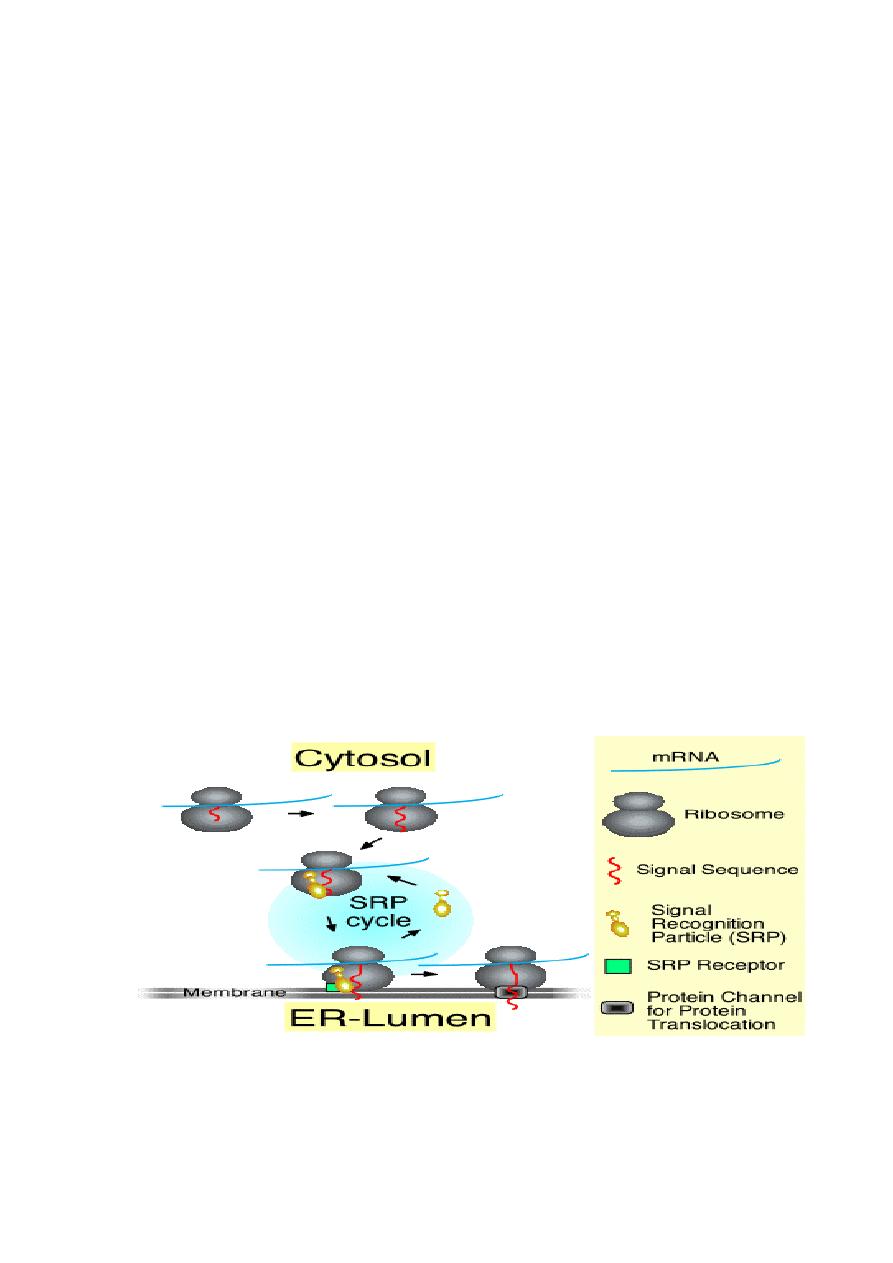
The SRP cycle
The signal recognition particle (SRP) associates with ribosomes
that are in the process of translating the mRNA for a secretory
protein. The protein has a signal at the N-terminus. Subsequently,
the ribosome-bound SRP interacts with the SRP-receptor a
component of the ER membrane. Finally, SRP recycles to
associate with another ribosome, and translation continues with
the secretory protein transversing the membrane through a
channel called the translocon.
Lecture 6: Protein targeting and degradation
Prof. Dr. Hedef Dhafir El-Yassin 2013
5
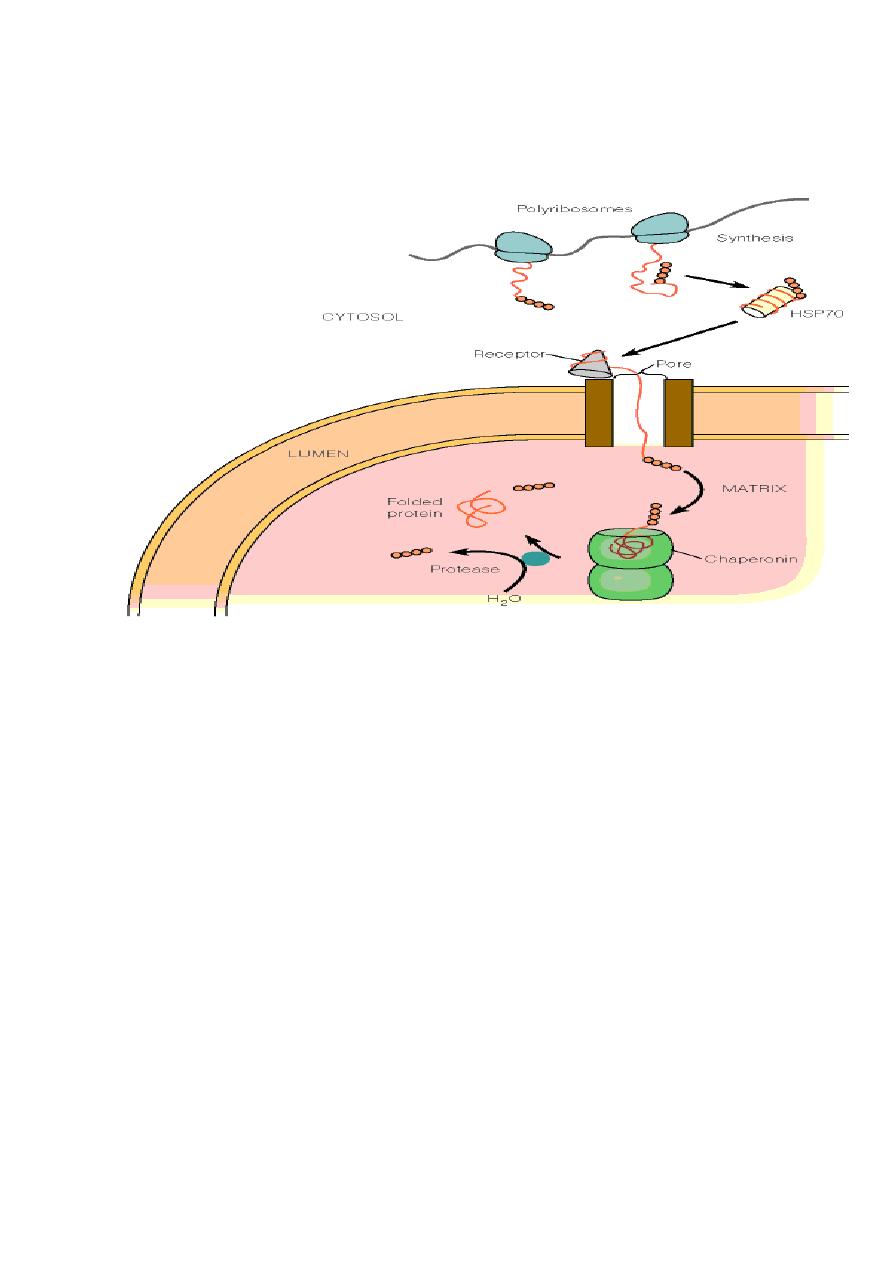
Proteins that must be targeted to the nucleus have a
nuclear
localization signal
(
NLS
). Once common type of signal is a series
of five or so closely spaced positively charged amino acids.
Protein Turnover
Protein lifetimes must also be regulated. Some proteins are
needed for only very short times -- and could be harmful is present
for too long. Others are needed all the time and it would be
unnecessarily wasteful to keep re-synthesizing them.
The following table, demonstrates the great differences in lifetimes
of some rat liver enzymes:

Lecture 6: Protein targeting and degradation
Prof. Dr. Hedef Dhafir El-Yassin 2013
6
Enzyme
Half-life
(h)
Ornithine decarboxylase
0.2
RNA polymerase I
1.3
Tyrosine
aminotransferase
2.0
Serine dehydratase
4.0
PEP carboxylase
5.0
Aldolase
118
GAPDH
130
cytochrome c
150
The lifetime of proteins in eukaryotes appears to be determined by
the nature of the N-terminal amino acid. Some amino acids (e.g.
Ala, Cys, Gly, Met, Pro, Ser, Thr, Val) stabilize proteins (at least in
yeast); others (e.g. Arg, His, Ile, Leu, Lys, Phe, Trp, Tyr)
destabilize proteins.

Lecture 6: Protein targeting and degradation
Prof. Dr. Hedef Dhafir El-Yassin 2013
7
Targeted Protein Degradation
In order to keep a cell working it needs to remove:
1. incorrectly synthesized proteins (with errors in amino acid
sequence)
2. damaged proteins (i.e. oxidative damage)
3. cell-cycle specific proteins
4. other signaling proteins which are no longer necessary
One mechanism of protein degradation is via lysosomes.
Lysosomes are acidic vesicles that contain about 50 different
enzymes involved in degradation:
1. proteases (cathepsins): cleave peptide bonds
2. phosphatases: remove covalently bound phosphates
3. nucleases: cleave DNA/RNA
4. lipases: cleave lipid molecules
5. carbohydrate-cleaving enzymes: remove covalently bound
sugars from glycoproteins
Lysosomes often secrete their contents into the extracellular
medium via exocytosis.
Lysosomes can also target damaged organelles in a process
called autophagy.
Sometimes, lysosomes are triggered to rupture inside a cell,
resulting in autolysis, also called apoptosis or programmed cell
death.

Lecture 6: Protein targeting and degradation
Prof. Dr. Hedef Dhafir El-Yassin 2013
8
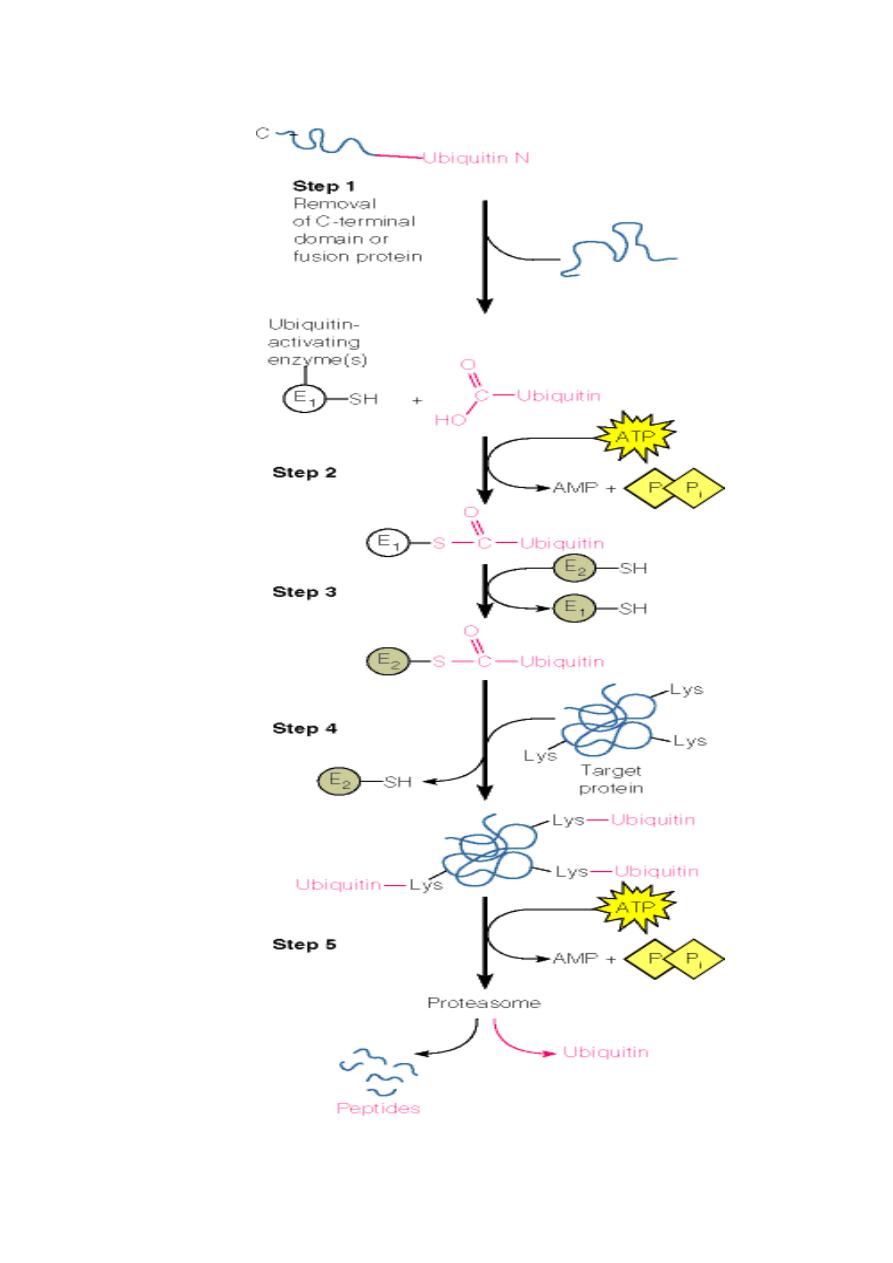
Another major mechanism is via ubiquitin labeling of surplus
proteins:
Ubiquitin (a small 76-residue protein) is attached to the
protein:
o
First, an activating enzyme attaches itself to the
carboxy terminus of free ubiquitin in an ATP-dependent
process.
o
Then, the activated ubiquitin is transferred onto a
second enzyme which at the same time recognizes
damaged proteins.
o
The activated ubiquitin is then covalently linked to
lysine residues on the surface of the damaged protein.
These ubiquitin-tagged proteins are now recognized by
specific proteases in the cytosol which in turn cleave and
degrade the tagged protein.
These proteases are combined in a very large protein
complex called the proteasome.
Lecture 6: Protein targeting and degradation
Prof. Dr. Hedef Dhafir El-Yassin 2013
9

Lecture 6: Protein targeting and degradation
Prof. Dr. Hedef Dhafir El-Yassin 2013
11
Conclusions:
1. Proteins synthesized on cytosolic ribosomes are released
into the cytosol or transported into mitochondria,
peroxisomes, and nucleus. Proteins synthesized on
ribosomes attached to the rough endoplasmic reticulum
(RER) are destined for lysosomes, cell membranes, or
secretion from the cell. These proteins are transferred to the
Golgi complex, where they are modified and targeted to
their ultimate locations.
2. In order to keep a cell working it needs to remove unwanted
protein
